Vol 8 No 4 2023 – 51
First report of Alternaria alternata associated with spot blotch of barley (Hordeum vulgare) in Iraq.
Itlal AlMasoodi 1 , Zainab Hameed 1 , Adnan Lahuf 2,*
1 Department of Field Crops/Agriculture College/ University of Kerbala/ Karbala/ Iraq; zainab.l@uokerbala.edu.iq.
2 Department of Plant Protection/Agriculture College/ University of Kerbala/ Karbala/ Iraq; adnan.lahuf@uokerbala.edu.iq..
* Correspondence: adnan.lahuf@uokerbala.edu.iq; Tel.: (008647810541064)
Available from. http://dx.doi.org/10.21931/RB/2023.08.04.56
ABSTRACT
A field survey was conducted during the barley growing season 2019 in the Karbala Province of Iraq. Barley plants with severe leaf spot symptoms were collected from several barley fields. The associated fungus was isolated and purified from those symptomatic leaves. The fungus colonies were aeriform mycelia in grey to black color. Conidia observed that they were light to dark brown and formed different-length chains. The causal agent was confirmed as Altenaria alternata via the PCR amplification and sequencing of the rDNA-ITS region, actA, and tef1 genes. Pathogenicity test showed that A. alternataisolated was pathogenic by producing light to dark brown spots on barley foliage. This is the first record of this pathogen causing Alternaria spot blotch on barley crops in Iraq.
Keywords: Alternaria alternata; leaf spot; Hordeum vulgare; Pathogenicity assessment; Phylogeny analysis.
INTRODUCTION
The Barley (Hordeum vulgare L.) is the fourth most common cereal crop after maize, rice, and wheat in the grass family Poaceae, 1. It is currently grown worldwide and can be grown in summer and winter in some temperate and subtropical regions; 2. Barley production worldwide in 2019 amounted to 142,910,932 tons, of which the Russian Federation continues to be the world’s leading producer, 11,88 %, then France, at 7,83% and Germany at 6,70%. However, Iraq produced only 571,283 tons, representing 0.39% of global production3. Besides, the average production per hectare was low at 16062kgg/ha compared to the average of 67930.4kgg/ha in the top 10 producers, 3.
Various abiotic and biotic stresses, including fungal diseases, can lead to a significant loss in different plant hosts, including barley crops, 4. Alternaria alternata (Fr.) Keissl is one of those fungi that infect a broad range of hosts, including barley, and causes different diseases. For instance, it was reported to cause leaf spots on Houttuynia cordata, 5, Ipomoea purpurea, 6 and apricot, 7. Furthermore, it was recorded infecting potatoes, lily and Catharanthus roseus, causing leaf blight, 8-10.
The Alternaria genus comprises 299 species with a significant morphological variety, 11. Conventionally, the identification of these species is based on their cultural and morphological characterizations, 12. However, this identification is complex and needs to be improved for most Alternaria species 12. Thus, these morphological descriptions should be combined with molecular techniques for obtaining accurate and price identification 13,14. Therefore, multiple DNA barcode genetic markers have been employed to detect, identify, and classify fungi comprising Alternaria species. For example, the internal transcribed spacer (ITS) region is a standard universal barcode applied successfully for fungal taxonomic differentiation and identification (add a reference). As well as the translation elongation factor 1-alpha (tef1), actin (actA), and DNA-directed RNA polymerase II (rpb2) genes are also frequently utilized for the same purpose, 15. However, occasionally, for some species, a specific alternative barcode is needed for sufficient identification, 16.
In Iraq, the etiology of spot blotch disease on barley has been rarely explored. Thus, the present study aimed to identify the fungus associated with this disease in Karbala province/ Iraq based on its morphological and molecular attributes.
MATERIALS AND METHODS
The Fungal isolation and morphological identification
In 2019, leaf samples of different barley varieties with typical spot blotch symptoms (Figure 1) were collected to determine the related causal agent. These samples were gathered from several barley fields in Al-Hussainiya territory (latitude 32°40’25.5″ N and longitude 44°09’47.3 «E), Karbala Province, Iraq. The symptomatic leaves were cut into small pieces (1-1.5 cm long), their surfaces disinfected with 2% sodium hypochlorite solution for 2 minutes and rinsed three times with sterile distilled water. The sterilized leaf pieces were then placed on a water agar medium (WA) and incubated at 25+2 °C for three days. To obtain pure colonies, the hyphal tip of each colony that emerged was transferred to Petri dishes containing potato dextrose agar media (PDA) and incubated at 25+2 °C. After one week, the morphological characteristics of the fungal isolates were examined through microscopic observations using a HumaScope PremiumLED compound microscope with a Digital Camera 5 Megapixel attached (HUMAN Gesellschaft für Biochemica und Diagnostica Germany). The average length and width of conidia were obtained by random measurement of 50 conidia, 17,18.
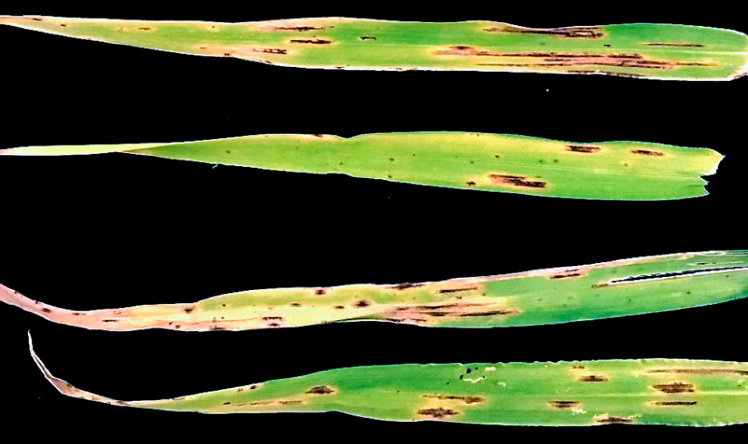
Figure 1. Typical symptoms of spot blotch on barley leaf samples.
Molecular characterization
The genomic DNA of the pure mycelia of seven-day-old colonies was extracted based on a developed protocol, 19. The quality and quantity of DNA extracted were evaluated via NanoDrop™ 2000/2000c Spectrophotometers device (Thermo Fisher Scientific, Waltham, MA). Subsequently, Polymerase chain reaction (PCR) test was executed using the internal transcribed spacer regions (ITS) primers, 20 (ITS1: 5`-TCCGTAGGTGAACCTGCGG-3` and ITS4: 5`-TCCTCCGCTTAT TGATATGC-3`), partial actin (actA) gene primers,21 (ACT-512F: 5`-ATGTGCAAGGCCGGTTTCGC-`3 and ACT-783R: 5`-TACGAGTCCTTCTGGCCCAT-`3), and partial translation elongation factor 1-alpha (tef1) gene primers 21(EF1-728F’5-CATCGAGAAGTTCGAGAAGG-‘3 and EF1-986R:`5-TACTTGAAGGAACCCTTAC C-`3). The PCR amplicons of ITS, actA and tef1 were sequenced employing their primers at Macrogen Inc. (Seoul, Korea; http://www.macrogen.com). Sequences were then edited via BioEdit version 7.0.5.3 and 22and compared by the nucleotide BLAST (www.ncbi.nlm.nih.gov) software with the GenBank database sequences. Phylogenetic analyses were conducted in MEGA-X, 23 using the maximum-likelihood model for the obtained ITS, actA and tef1 sequenced, 24.
Pathogenicity assessment
The pathogenic potential of fungal isolates was verified on barley leaves of Al-Warkaa cv. Utilizing the detached leaf assay, 25. A conidial suspension at a concentration of 2000 conidia per ml was prepared from 7-day-old pure cultures. A 50-μl of this suspension was applied onto the adaxial side of each healthy leaf (n=12) that was surface disinfected with 70% ethanol. Other disinfected leaves were treated with distilled water only for control purposes. The treated leaves were then sited on moist tissues into plastic containers that were incubated at 25+2 °C in darkness and periodically observed until disease symptoms appeared. The fungal pathogen was re-isolated from spots blotch of inoculated leaves, and its morphological features were re-examined to complete Koch’s postulates. The test was repeated twice to confirm the results obtained, 26.
RESULTS
Morphological identification
A total of 20 fungal isolates were obtained consistently from the diseased barley leaves. However, no apparent differences in cultural or morphological characterizations were noticed among these isolates. At the beginning of the fungal development, the colony’s growth on PDA was abundant airy mycelium in white-greyish color with light to dark green in internal areas extending from the colony’s center. Laterally, the mycelium color converted to dark brown or dark olivaceous bounded with light grey mycelium. However, the colonies were dark brown to black on the reverse surface.
Furthermore, the microscopic analysis showed that the conidiophores were typically branched or simple, usually curved with a length of 12 to 15 μm and a width of 4 to 6 μm. Conidia (Figure 2) were pale to light brown and typically formed in branched chains of different shapes. They were generally ovoid, obclavate or ellipsoidal shape with an average length of 22–30 μm and a width of 16–18 μm developing a short conical beak at their tips and owned 2-5 transverse septa and 1-2 longitudinal septa. According to these characteristics, which agreed with the previous description12, the fungus was initially identified as A. alternata.
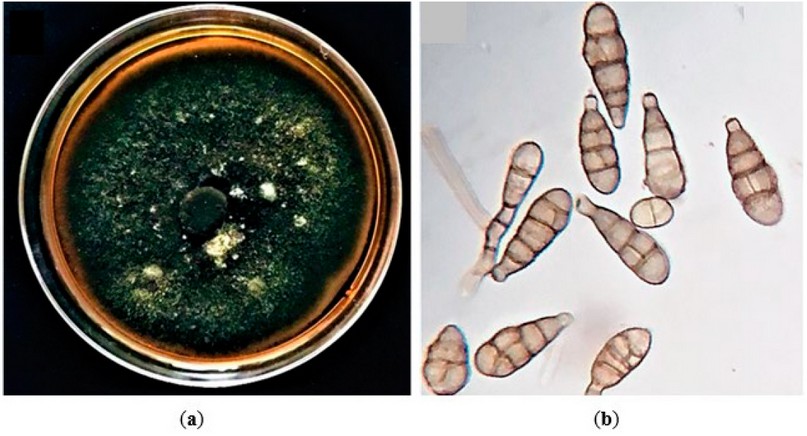
Figure 2. The culture and morphology of A. alternata were isolated in this study. (a) Colony morphology of the representative isolate, Alhusseiniya, cultured on PDA; (b) Microscopic observation of the A. alternata conidia.
Molecular characterization
The initial identification of the A. alternata isolated was confirmed through successful amplification and sequencing generated for the ITS region, tef1 and actA genes, respectively. The BLAST analysis revealed 99-100% similarity of the sequences obtained in this study with those of numerous A. alternata isolates deposited in the GenBank database, as the phylogenetic analysis confirmed the identification of this species by clustering it with other references of A. alternata in the three genetic markers (Figure. 3,4,5). Accordingly, they were deposited in the GenBank and had specific accession numbers (ITS-MT874979.1; TEF1-MT875391.1; actA-MT875390.1).
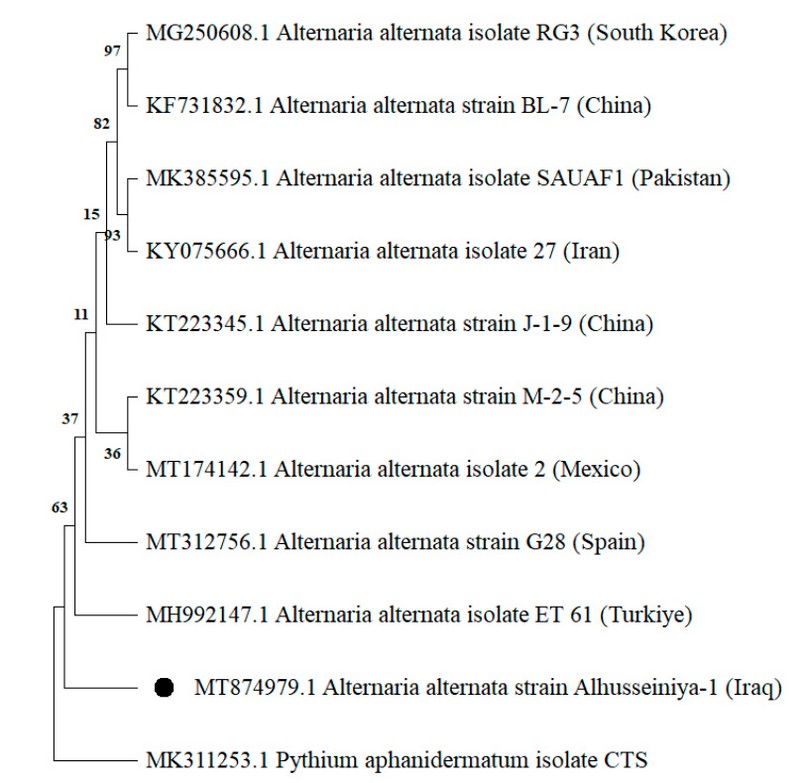
Figure 3. Phylogenetic tree created by neighbor-joining process gathered from ITS-rDNA region sequences presenting the relationships among isolates of A. alternata isolate Alhusseiniya-1 (indicated with a black dot) and related Alternaria species. The out-group fungus is Pythium aphanidermatum.

Figure 4. The phylogenetic tree produced by the neighbor-joining procedure collected from tef1 gene sequences displays the relationships among isolates of A. alternata isolate Alhusseiniya-1 (indicated with a black dot) and related Alternaria species. The out-group fungus is Anthanassa drusilla.
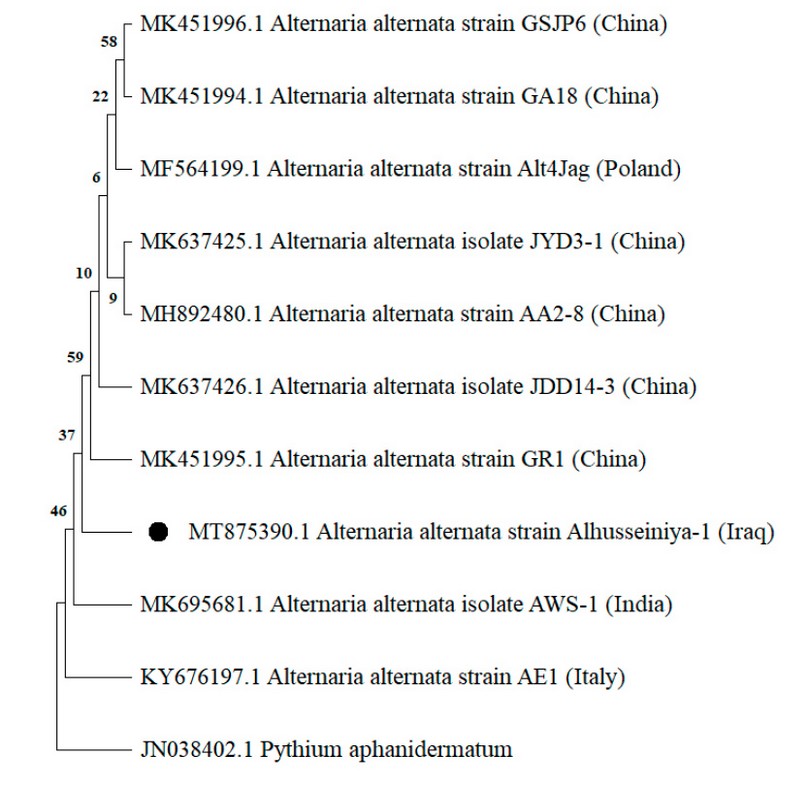
Figure 5. Phylogenetic tree constructed by neighbor-joining procedure collected from actA gene sequences showing the relationships among isolates of A. alternata isolate Alhusseiniya-1 (indicated with a black dot) and related Alternaria species. The out-group fungus is Pythium aphanidermatum.
Pathogenicity assessment
The A. alternata isolate Alhusseiniya tested was pathogenic, producing symptoms on the detached leaves (Figure 6) starting after about one week. The typical symptoms initially appeared as small brown necrotic lesions that subsequently extended to be more prominent in oval shapes with distinct chlorotic or straw-color edges. These lesions ultimately combined and caused the drying of leaves. However, control leaves did not develop any disease symptoms. The fungal pathogen was re-isolated from the symptomatic barley leaves, and the microscopic observation demonstrated that its morphological features were similar to those of the initially obtained isolate.
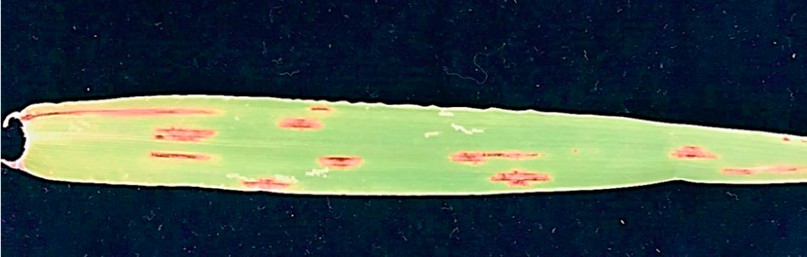
Figure 6. Pathogenicity test on barley leaf. The inoculated leaf showed typical symptoms after 14 days post-inoculation.
DISCUSSION
In this study, A. alternata isolate Alhusseiniya isolated from barley leaves was identified based on its morphological, molecular, and pathogenicity characteristics. A. alternata is supposed to be worldwide distributed, causing various plant diseases such as root rots, blights and leaf spots on a wide range of host plants, 5-10. To distinguish Alternaria spp., morphological characters comprising sizes and shapes of conidia and chain formation are categorized the genus into 14 species, 12. However, diverse morphologic flexibility has been observed among Alternaria spp due to the influence of numerous factors such as temperature, light and humidity. As well as, an extensive morphological variation of septation number, shape, color, and size of conidia was based on differentiation in conidium age, 12. Therefore, identifying Alternaria species based on their morphological features is complex, insufficient, and inaccurate. Due to these limitations, several molecular methods were suggested to be applied besides morphological characterizations for accurately identifying Alternaria species and other microorganisms, 13, 23.
The sequencing of numerous universal genetic barcodes besides morphological traits has been used successfully in identifying Alternaria spp—pathogens, including A. alternata. For example, A. alternata causing leaf spots on quince trees and Rehmannia glutinosa plants was identified relying on the morphological characterizations and sequencing of the translation elongation factor 1-alpha (tef1), internal transcribed spacer (ITS), actin (actA), glyceraldehyde-3-phosphate dehydrogenase, RNA polymerase II second largest subunit, major allergen Alt a 1 gene, endopolygalacturonase gene and anonymous gene region, 27. The sequences of the Internal transcribed spacer and β-tubulin were utilized in identifying the same species causing fruit Rot of opium poppy and black spots on pink lapacho, 28.
The pathogenicity assessment proved that A. alternata isolates Alhusseiniya were pathogenic to this crop by producing varying spot blotch symptoms. The disease incidence was estimated to be around 20-30% in all barley fields surveyed in the Alhusseiniya region of Karbala province, where the disease was noticed. However, previous studies have recorded A. alternata associated with different diseases on various crops, 29. It has been reported only as one of the causal agents of spot blotch disease on barley crops, 30. However, this research’s findings and prior reports suggest the polyphagous aspect of this species and its adaptation to more significant geographic regions. The leaf spot blotch disease of barley caused by A. alternata is occurring in Karbala province and possibly in barley-producing regions of Iraq. However, it was undetected by the farmers until it was reported in the present study. Thus, this is the first record of A. alternata causing spot blotch on barley in Iraq. This discovery is highly significant regarding the fungal epidemiology and management of the disease using new control and management methods that were proved to be efficient against various plant pathogens such as biological agents and nanoparticles, 31-33.
CONCLUSIONS
In this study, the etiology of spot blotch disease on barley crops has been identified as the fungus A. alternata for the first time in Iraq. This identification was based on the fungal pathogen’s morphological, molecular and pathogenicity characteristics.
Author Contributions: Conceptualization and methodology, A. L.; investigation, Z. H.; resources and writing-original draft preparation, I. A.; writing—review and editing, Z. H.; visualization, I. A. and Z. H.; supervision and project administration, A. L. All authors have read and agreed to the published version of the manuscript.
Funding: This research received no external funding.
Data Availability Statement: Data supporting these reported results can be found at https://www.ncbi.nlm.nih.gov/nuccore/MT874979.1; https://www.ncbi.nlm.nih.gov/nuccore/MT875391.1; https://www.ncbi.nlm.nih.gov/nuccore/MT875390.1
Acknowledgments: The authors would like to acknowledge the administrative support by the Department of Plant Protection, Agriculture College, and the University of Kerbala.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1. Stein, N. ; Muehlbauer, G. J. The Barley Genome; Springer International Publishing: Gewerbestrasse, Switzerland, 2018;pp.1-430.
2. Ullrich, S. E. Barley production, improvement, and uses; John Wiley & Sons Ltd: West Sussex, UK,2011; pp. 1-300.
3. FAO. Food and Agriculture Organization. The database of annual production. FAOSTAT. Statistical database. Available online: http://www.fao.org/faostat/en/#data /QC. (accessed on 25 September 2021).
4. Lah.uf, A. A. First report of Fusarium proliferatum causing stem and root rot on lucky bamboo (Dracaena braunii) in Iraq. Hell. Plant Prot. J. 2019, 12 (1),1-5. https://doi.org/10.2478/hppj-2019-0001.
5. Zheng, L. ; Lv, R. ; Li, Q.; Huang, J.; Wang, Y.; Hsiang, T. First report of leaf spot of houttuynia cordata caused by Alternaria alternata in China. Plant Dis. 2011, 95, 3. https://doi.org/10.1094/PDIS-10- 10-0724.
6. Yang, Q. Q. ; Zhou, S. Y. ; Liang, W. X.; Li, L. First Report of Alternaria alternata causing leaf spot on Ipomoea purpurea in China. Plant Dis. 2017, 102, 2. https://doi.org/10.1094/PDIS-08-17-1278-PDN.
7. Hameed, Z. L.; Lahuf, A. A. ; Jasim, M. T.; Mohsen, H. M. ; Kadim, B. J. ; Saleh, S. A. ; Mohamed, A.F. First Report of Alternaria Alternata causing brown leaf spot on apricot (Prunus Armeniaca) in Karbala Province of Iraq. Earth Environ. Sci. 2021, 910, 012080. https://doi.org/10.1088/1755-1315/910/1/ 012080.
8. Van Der Waals, J.E.; Pitsi, B.E.; Marais, C.; Wairuri, C.K. First report of Alternaria alternatacausing leaf blight of potatoes in South Africa. Plant Dis.2011, 95, 363-363. https://doi.org/10.1094/PDIS-11-10-0820.
9. La,huf, A. A.; Abdalmoohsin, R. A. G.; Alhusani, A. H.; Al-Asadi, A. First report of leaf blight disease in lily (Lilium candidum) caused by Alternaria alternata in Iraq. Biopestic. Int. 2018, 14 (2), 123-126.
10. Lahuf, A. A. Alternaria alternata causes leaf blight of rosy periwinkle (Catharanthus roseus) in Iraq. Aust. Plant Dis. Not. 2019, 14:4.https://doi.org/10.1007/s13314-019-0334-9.
11. Lahuf, A. A.; Abdullah, K. M.; Mohammadali, M. T. Assessment of the nanosized particles of ZnO and MgO and some cultivars in control of Alternaria solani causing tomato early blight. Ecol. Environ. Conserv. 2020, 26, 89-95.
12. Simmons, E.G. Alternaria. An Identification Manual, CBS Biodiversity Series 6.; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. pp.22-39.
13. Shehan, S.J.; Abdalmoohsin, R.G.; Lahuf, A.A. First report of Ectophoma multirostrata causing root rot of Celosia argentea in Iraq. New Dis. Rep. 2022, 46, e12126. https://doi.org/10.1002/ndr2.12126.
14. Bajinka, O. ; Secka, O. Integration of Molecular Methods into Microbiological Diagnostics. Appli. Micro. 2017, 3, 130.
15. Noaman A I, Khalaf R M, Emad GH, Al-Abbasy, Mohammed Th. T. Effect of flaxseed oil dosing on fertility, growth characteristics and some physical, biochemical, and hormonal blood parameters during the early pregnancy of Awassi ewes. Revis Bionatura. 2022;7(4) 5. http://dx.doi.org/10.21931/RB/2022.07.04.5.
16. Jaber, M.H.; Lahuf, A.A. Survey, pathogenicity and molecular identification of novel Fusarium species causing seed decay and damping-off disease of wheat crop in Kerbala Province, Iraq. Plant Cell Biotechnol. Mol. Biol. 2020 21, 1–14.
17. AL-Musawi, M. A. ; Lahuf, A. A.; Jafar, O. H . Isolation and diagnosis of the pathogens causing seed decay and damping-off disease on wheat and control them using some biological and chemical factors. Journal of Kerbala for Agricultural Sciences 2017, 4 (1), 113-132. https://doi.org/10.59658/jkas.v4i1.88.
18. Lahuf, A. A.; Jaafar, O. H.; Al-mosoy, M.; Hameed, Z. L. First record of the crown rot fungus Fusarium equiseti affecting Triticum aestivum L. and Aptenia cordifolia in Iraq. Asian J. Agric. Biol. 2018, 6 (4), 543-548.
19. Sh. Al-Obaidy, G., F. Abdulrahman, M., Sh. J. Alobaidy, B. Future Trends For Green Environmental Applications Of Nanotechnology: A Review. Anbar Journal Of Agricultural Sciences, 2023; 21(1): 133-147. doi: 10.32649/ajas.2023.179725.
20. White, T.; Bruns, T. ; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocol: a guide to methods and applications. Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, USA, 1990.pp.22-34.
21. Al-Tememe, Z.A.M.; L.ahuf, A.; Abdalmoohsin, R.G.; Al-Amirry, A.T. Occurrence, identification, pathogenicity and control of Neoscytalidium dimidiatum fungus, the causal agent of sooty canker on Eucalyptus camaldulensis in Kerbala Province of Iraq. Plant Arch. 2019, 19, 31–38.
22. Hall, T.A., BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. No. 41. Nucleic Acids Symposium Series 1999, 41, 95–98.
23. Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 2015, 33, 1870–1874. https://doi.org/10.1093/molbev/msw054.
24. Zain, H.; Tatar , A.; Alabi, O. M. .; Samiei Zafarghandi, M. . The Effect Of Using Different Levels Of Vitamin E On The Antioxidants Status Of Broiler Chickens. JLSAR 2023, 4, 37-44.
25. Akhtar, K.P.; Sarwar, N.; Saleem, M.Y.; Asghar, M. Convolvulus arvensis, a new host for Alternaria solani causing early blight of Solanum lycopersicum in Pakistan. Aust. Plant Dis. Not. 2011, 6(1), 84–86. https://doi.org/10.1007/s13314-011-0029-3.
26. Khudai M Y, Abdulateef S M, Mouhammed T Th, Alamili H S. Use of modern geometric design of fish ponds to increase welfare and blood parameters. Revis Bionatura 2023;8 (2) 82. http://dx.doi.org/10.21931/RB/2023.08.02.82.
27. Guo, L.; He, S.; Gao, Z.; Wu, Z.; Duan, R.; Yang, K.; He, X. First report of fruit rot in opium poppy (Papaver somniferum L.) caused by Alternaria alternata in China. Plant Dis. 2020, 104, 3264-3264. https://doi.org/10.1094/PDIS-01-20-0211-PDN.
28. Ali, H.H., AL-Rawi, K., Khalaf, Y., Alaaraji, S., Aldahham, B., Awad, M., Al-ani, O., Al-ani, F., Ali, A.T. Serum Caveolin-1 Level is Inversely Associated with Serum Vaspin, Visfatin, and HbA1c in Newly Diagnosed Men with Type-2 Diabetes (2022) Reports of Biochemistry and Molecular Biology, 11 (2), pp. 299-309.
29. Ramakant, R.; Joshi, P.; Bhandari, M.S.; Pandey, A.; Pandey, S. Identification and pathogenicity of Alternaria alternata causing leaf spot and blight disease of Ailanthus excelsa in India. For. Pathol. 2019, 00:e12584. https://doi.org/10.1111/efp.12584.
30. Aada, A. M. M. A. Identification of pathogens and control of spot blotch disease of barley (Hordeum vulgare) by combining plant resistance and biological control. A PhD. Thesis, Newcastle University, Newcastle, UK. 2013.
31. Lahu.f, AA; Kareem, A.A.; Al-Sweedi, T.M.; Alfarttoosi, H.A. Evaluation the potential of indigenous biocontrol agent Trichoderma harzianum and its interactive effect with nanosized ZnO particles against the sunflower damping-off pathogen, Rhizoctonia solani. IOP Conf. Ser.: Earth Environ. 2019, 365, 012011. https://doi.org/10.1088/1755-1315/365/1/012033
32. Lahuf, A.A.; Alfarttoosi, H.A.; Al-Sweedi, T.M.; Middlefell-Williams, J.E. Evaluation of an integration between the nanosized zinc oxide and two cultivars for the control of damping-off disease in sunflower crop. Res. Crops. 2019, 20,1,174–179. https://doi.org/10.31830/2348-7542.2019.024.
33. Abdulmoohsin, R. G.; Lah,uf, A. A.; Husain, Y. N.; Hameed, Z. L. Bioefficiency of some indigenous biocontrol agents against Rhizoctonia solani causing cowpea seed rot and preemergence damping-off. IOP Conf. Ser.: Earth Environ. Sci. 2019, 388, 012011. https://doi.org/doi:10.1088/1755-1315/388/1/ 012011.
Received: 26 September 2023 / Accepted: 15 April 2023 / Published:15 December 2023
Citation: AlMasoodi, I; Hameed, Z; Lahuf, A. First report of Alternaria alternata associated with spot blotch of barley (Hordeum vulgare) in Iraq. Revis Bionatura 2023;8 (4) 56. http://dx.doi.org/10.21931/RB/2023.08.04.56



















