Vol 8 No 2 2023 – 91

Isolation and diagnosis of some associated fungi with cowpea root rot disease and testing its pathogenicity
Ahmed Jassim Awad1 and Theyab A.Farhan1
1 Department of Plant Protection, College of Agriculture, University of Anbar, Anbar, Iraq; ahm20g7001@uoanbar.edu.iq
* Corresponding author, deab.frahen@uoanbar.edu.iq;
Available from: http://dx.doi.org/10.21931/RB/2023.08.02.91
ABSTRACT
Execute search by date 1/4/2021, The results of collecting samples from the regions of Anbar, Baghdad, Salah al-Din and Wasit showed that cowpea root rot disease is widespread in all studied areas, and The results of isolation phenotypic and molecular diagnosis showed the presence of different isolation of fungi that infected cowpea root, such as Fusarium nygamai (Fu1), F. nygamai (Fu2), F. solani (Fu3), F. solani(Fu4), Rhizoctonia solani (Rh5), and Fusarium solani (Fu6), The results of the pathogenicity test on radish seeds showed that tested isolates were significantly decreased germination percentage of radish seeds of water Ager, and the most effective isolation was F. solani (Fu4). The infection rate was 90% compared to 0.00% of the control media treatment, which was uncontaminated by the pathogenic fungus. Isolated fungi showed a difference in the percentage and severity of infection on cowpea seedlings and seedlings, as the isolate of F.nygamai (Fu1) achieved the highest infection rate of 66.67 % and the severity of disease at 75%. All fungal isolates significantly increased the rate and severity of infection on seed radish compared with the control treatment not contaminated with pathogenic fungi by 0.0%.
Keywords: Vegan unguiculata; Rhizoctonia solani; and Fusarium solani; PCR.
INTRODUCTION
Cowpea (Vegan unguiculata) is a multi-purpose crop belonging to the legume family, and it is grown in dry and semi-dry tropical regions1. The global cowpea production is estimated at 4.5-6.5 million tons, and 80% of production is from Africa,2. In Iraq, the cultivated area of cowpeas reached 72,507 m2, and the output of one dunam ranged between 2600-2768 kg,3. Copwea is affected by many diseases, such as root rot and damping off, as these diseases cause losses that may reach 55% of the cowpea production,4–5. Moreover, the losses can get 100% yield in the appropriate environmental conditions,6–7. Root and stem rot disease is caused by fungi such as Fusarium solani and Rhizoctonia solani, the most dangerous fungi in cowpea, 8-9, because they contain toxins and enzymes (fludarabine, fusaric acid, javanicine, pectin methyl esterase and galacturonic enzymes), as well as their ability to tolerate toxins and antibiotics secreted by other organisms, 10-11. Fusarium solani and Rhizoctonia solani fungi are classified based on their phenotypic characteristics,12,13-14. However, several studies have been using Polymerase Chain Reaction technology (PCR) as a modern application in diagnosing fungi based on the nitrogenous bases sequence traits in a single DNA strand, 15, 16–17. Found 18-19 that the root rot disease and damping off of cowpea seedlings, caused by Fusarium solani and Rhizoctonia solani, have caused significant losses in yield, and they are the most common pathogens that infect cowpea.
MATERIALS AND METHODS
Sample collection
Samples of infected cowpea plants by fungi were collected from different governorates of Iraq, Anbar, Baghdad and Wasit and Salah Al-Din, in June, July, August and September 2021. that showed symptoms of root rot diseases were collected, such as leaves wilting, burning of the leaves edges, discoloration of the roots and plant death,20.
Isolation of associated fungi with cowpea root rot disease
Infected parts of cowpea plants were cut into small pieces 0.5-1 cm in length and superficially sterilized using a solution of 6% sodium hypochlorite (Naocl) free chlorine, then washed with sterilized distilled water to remove the remnants of the sterilizing solution and placed on a filter paper, then transferred into Petri dishes 9 cm containing Potato Dextrose Agar (PDA) and incubated at 25 ± 2 °C for 5-7 days until a formation of the fungal growth, the growing fungi were purified by taking a small part of the edge of The fungal growths, and transferred in the center of another Petri dishes containing (PDA) average of three replicates then incubated at a temperature of 25 ± 2 °C.
Phenotypic diagnosis
Fungi isolates were diagnosed after purification of the growth of fungal colonies was completed on the PDA medium., Microscopic slides were prepared and checked using a light microscope; the fungi were diagnosed to the genus level depending on the phenotypic characteristics by using the approved taxonomic keys,21-22.
Molecular Diagnosis
DNA of six isolates, F. nygamai (Fu1), F. nygamai (Fu2), F. solani (Fu3), F. solani (Fu4), R. solani (Rh5) and F. solani (Fu6), Scientific Progress Laboratory – Baghdad- Al-Harithiya, was extracted by using standard kit ABI OPure from the American company., The extraction was performed in the Scientific Progress Laboratory, located in Baghdad,/Iraq, and diagnosis steps were described by 23. The polymerase chain reaction (PCR) was prepared according to 24 and the primersITS1.5′-TCCGTAGGTGAACCTGCGG-3.and.ITS4 5′-TCCTCCGCTTATTGATATGC-3(Macrogen Company, Korea) were used. The lyophilized primers were dissolved in nuclease-free water to give a final concentration of 100 μl. After completing the DNA amplification and migration steps on agarose gel, the samples were sent to the Korean Macrogen Company to obtain Sequencing (Nitrogenous bases sequence).
Pathogenicity test on radish seeds
The fungus Fusarium spp and Rhizoctonia solani isolations were tested on radish seed in the Plant Protection Department, /College of Agriculture, /Anbar University, Iraq. The test of pathogenicity was carried out according to the method of 25. Data were statistically analyzed according to the completely randomized design (CRD), and the germination percentage of radish seeds average of three replicates calculated after 14 days of sowing seeds on PDA medium was calculated and compared with the control treatment them by following the equation below: Germination percentage = (number of germinated seeds)/(total seed number) × 100
Pathogenicity test on cowpea seed and seedling
Inoculum of fungal isolates was prepared according to the26 by using millet seeds, the soil was sterilized with commercial formalin, then sterilized soil was placed in sterilized plastic pots with a capacity of 2 kg, and pathogenic fungi inoculum was added at a rate of 1% (weight-weight), the process was repeated average of three replicates on all fungal isolates, and then the pot was rinsed with water and covered with polyethylene bags for 3 days after that cowpea seeds were planted at a rate of three seeds per pot, the sterilized cowpea seeds were placed without adding the pathogenic fungus inoculum as the control treatment, three replicates were used for each treatment. The test was carried out by using a CRD design, and the percentage of germination was calculated according to the following equation:
Germination percentage = (number of infected plants)/(total number of plants) x 100
The percentage of infected severity was calculated according to equation 27 as follows:
% Infected severity= ×100
0= no injury (healthy roots )
1 light brown coloration in the secondary roots
2 light brown coloration in the secondary roots, with a small part of the main root
3 dark brown coloration in the main roots, with no rotting of the stem bases
4 dark brown coloration of the roots, with rotting of the stem bases
5 plant death
RESULTS
Sample collection
The results in Table (1) showed that cowpea root rot diseases were widespread in all studied sample collection areas, which is consistent with what was mentioned by 28-9; the condition is dangerous and widespread in most countries where the crop is grown.
Phenotypic diagnosis
The results of the phenotypic diagnosis in Table (2) and Figure 1 showed the presence of fungi (Fusarium spp., and Rhizoctonia solani), which were associated with cowpea root rot disease, which is the leading cause of the disease,29-30.

Table 1. The infected sample of cowpea plants collection areas and the collection date
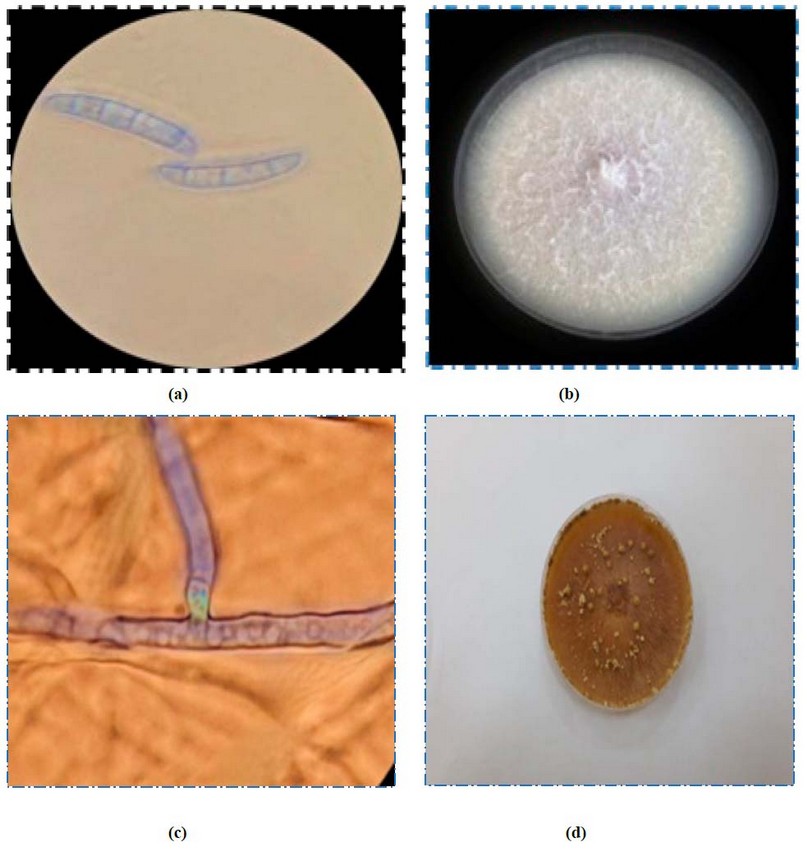
Figure 1. is the morphological and microscopic check of fungus colonies in the PDA medium.
a.b Fusarium solani and c. d Rhizoctonia solani
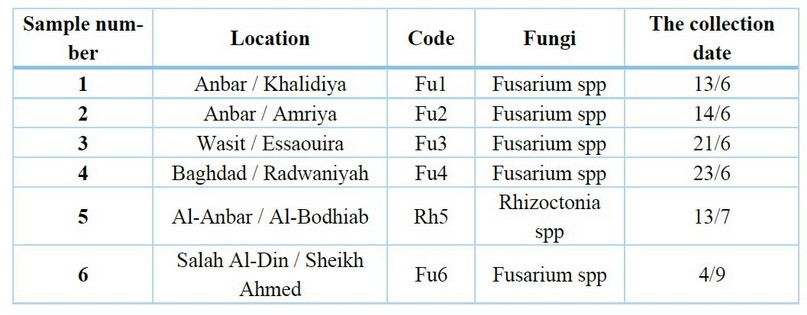
Table 2. The associated fungi with cowpea root rot disease were isolated and diagnosed phenotypically to the genus level
Molecular Diagnosis
The results of the molecular diagnosis by using the PCR technique in Table (3) and Figure (2), according to the arrangement of the nitrogen bases in the single DNA strand, showed that most types of fungi that cause cowpea root rot disease belong to Fusarium spp and Rhizoctonia solani. These results reinforce what was indicated by 34–35, that these fungi are the leading cause of the disease.
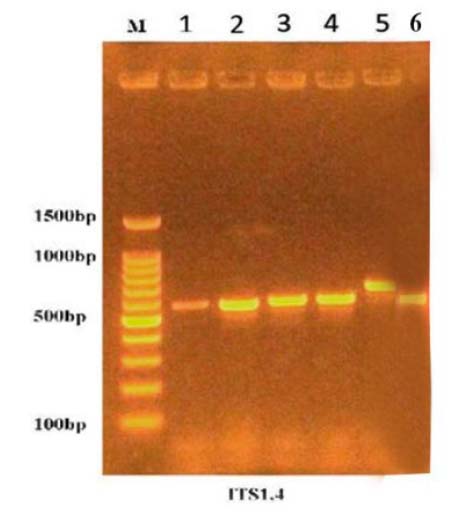
Figure 2. Amplification of ITS gene for unknown fungal species on agarose gel

Table 3. The associated fungi with cowpea root rot were diagnosed by using the PCR technique
Pathogenicity test on radish seeds
The results in Table (4) and Figure (3) showed that the pathogenicity test for some isolates of fungi (Fusarium nygamai (Fu1)- Khalidiya, Fusarium nygamai (Fu2)- Amriya, Fusarium solani (Fu3)- Essaouira, Fusarium solani (Fu4) Radwaniya, Rhizoctonia solani ( Rh5), Fusarium solani (Fu6), Fusarium solani (Fu7) achieved a significant reduction in the percentage of germination of radish seeds compared to the control treatment (untreated with pathogenic fungi) in which the infection rate was 0.0% 20, 36–37 or. 38–39
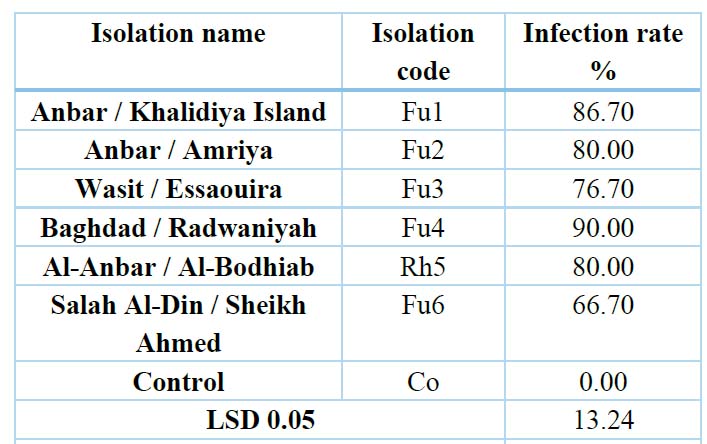
Table 4. Effect of some isolated fungi from cowpea roots on the germination of radish seeds on water agar medium

Figure 3. Effect of fungi isolates on the germination of radish seeds
Pathogenicity test on cowpea seed and seedling
The results of pathogenicity examination testing of cowpea seeds in Table (5) and Figure (4) showed that all the tested isolates (Fusarium solani, Fusarium nygamai, Fusarium nygamai, Fusarium solani, Fusarium solani, Rhizoctonia solani) varied in their impact on the ratio and severity of cowpea root rot disease, where, the isolate of Al-Anbar – Khalidiya Island (Fusarium nygamai) (Fu1) achieved the highest percentage and severity of infection, which amounted to 66.67% and 75%, respectively, and all tested isolates achieved a significant increase in the ratio and severity of disease compared to the control treatment (without adding pathogenic fungi), which reached 0.0% for both.

Table 5. Effect of some isolated fungi from cowpea roots on infecting the seeds and seedlings of cowpea under field conditions
Figure 4. Effect of isolate (Fu 4) Fusarium solani on seeds and seedlings of cowpea under field conditions
DISCUSSION
The phenotypic diagnosis of F. solani was made based on the shape of the large conidia (Macrioconidia), the presence or absence of small conidia, and the presence of Chlamydiospores whose location is terminal or intertwined, 31-32—F.solani based on the shape of the crescent-shaped Macroconida,33. As for the fungus Rhizoctonia solani, the thread is branched at right angles in the area of genesis with the presence of a constriction, and it is considered one of the diagnostic features of the fungus over other fungi,29. The difference in the effect of these isolates is due to their difference in the speed of penetration of plant cells and their ability to produce toxins and enzymes that break down plant cell walls; the results of this study agree 11.
CONCLUSIONS
The cowpea root rot disease spread in all the governorates from which the plant samples were collected. The leading cause of cowpea root rot disease is several species belonging to the genus Fusarium and the fungus Rhizoctonia solani—the difference in the pathogenicity of fungal isolates in causing cowpea root rot disease.
REFERENCES
1. Boukar, O., Belko, N. Chamarthi, S. Togola, A. Batieno, J. Owusu, E . Haruna, M. Diallo,S. Umar, M. L. Olufajo,O. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed 2019, 138, 415-424.
2. Jayathilake, C., R. Visvanathan, A. Deen, R. Bangamuwage, B.C. Jayawardana, S. Nammi, and Liyanage, R. Cowpea: An overview of its nutritional facts and health benefits. Journal of the Science of Food and Agriculture 2018, 98(13), 4793-4806.
3. Annual Statistical Collection. Agricultural Statistics Directorate. Central Statistical Organization. Ministry of Planning. Iraq 2020.
4. Alabouvette, C., Hoeper, H. Lemanceau, P. and Steinberg, C. Soil Suppressiveness to diseases induced by soil-borne plant pathogens.1996, P. 371 – 413.
5. Rodrigues ,A.A. Bezerra Neto, C.E. and Coelho R.S.B. induco de Resistencia Fusarium oxysporum f.sp Tracheiphilum Em caupi : Eficiencia de indutores Atioticos EAtividade Enzimatica Elicitada .Braz. phytopathol 2006. 31, 492-499.
6. Da Silva, V.B., Bomfim, C.S.G. Sena, P.T.S Santos, J.C.S., Mattos, W.D.S., Gava, C.A.T., and Fernandes-Júnior, P.I. Vigna spp. Root-Nodules Harbor Potentially Pathogenic Fungi Controlled By Co-habiting Bacteria. Current Microbiology 2021, 78(5), 1835-1845.
7. Graham, P.H., and Vance, C.P. Legumes : Importance and constraints to greater use. Plant physiology 2003,131,872-877
8. Lodha, SATISH, and Burman, U. Efficacy of composts on nitrogen fixation, dry root rot (Macrophomina phaseolina) intensity and yield of legumes. Indian Journal of Agricultural Science 2000, 70(12), 846-849.
9. Rauf, B.A. Seed-borne disease problems of legume crops in Pakistan. Pakistan J. Sci. and Indust. Res 2000, 43, 249-254.
10. Tewoldemedhin, Y.T., S. C. Lamprecht., M. M. Vaughan., G. Doehring and, O‘Donnell, K. Soybean SDS in South Africa is caused by Fusarium brasiliense and a novel undescribed Fusarium sp. Plant disease 2017, 101(1), 150-157.
11. Zheng, N., Zheng, L.P., GE, F.Y., Huang, W.K., Kong, L.A., Peng, D.L., and Liu, S.M. Conidia of one Fusarium solani isolate from a soybean-production field enable to be virulent to soybean and make soybean seedlings wilted. Journal of Integrative Agriculture 2018, 17(9), 2042-2053.
12. Kee, Y.J., Zakaria, L. and Mohd, MH Morphology phylogeny and pathogenicity of Fusarium species from Sansevieria trifasciata in Malaysia. Plant Pathology 2020, 69(3), 442-454.
13. Leslie, J.F. and Summerell . The Fusarium Laboratory Manual. Blackwell Publishing, Ames, IA. 2006; p .388.
14. Mc-Govern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Protection 2015,73,78-92.
15. Ezekiel, C.N., Oyedele, O.A. Kraak, B. Ayeni, K.I. Sulyok, M. Houbraken, J. and Krska, R. Fungal diversity and mycotoxins in low moisture content ready-to-eat foods in Nigeria. Frontiers in Microbiology 2020, 11, 615
16. Frisvad, J. C., Hubka, V. Ezekiel., C. N. Hong, S. B. Nováková, A. Chen, A.J. and Houbraken, J. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in mycology 2019, 91(1), 37-59.
17. Schoch, C.L., Seifert, K.A. Huhndorf, S. Robert, V. Spouge, J.L. Levesque C.A. and Miller A.N. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi .Proceedings of the National Academy of Sciences 2012, 109, 6241-6246.
18. Al-Moussawi, Mohsen Abd Ali Mohsen and Kamel Salman Jabr. Isolation and identification of the pathogen of cowpea root rot disease and evaluation of the efficacy of Azotobacter vineland in controlling the disease under laboratory conditions. Iraqi Journal of Agricultural Sciences 2012, 43(2), 67-75.
19. Wanas, Russell Ali. Isolation and identification of fungi accompanying the roots of cowpea (L.) Vigna ungniculata and integration in their control. Master Thesis. Faculty of Agriculture. University of Kufa 2011.
20. Agrios , G. N.. Plant Pathology. 5th Ed. Elsevier Inc. USA, 2005 ;P. 998.
21. Barnett, H. L., and Hunter, B. B. Illustrated genera of imperfect fungi. The American Phytopathological Society. US Department of Agriculture, Agricultural Research Service, Washington State University, Pullman. APS Press. USA. St. Paul, Minnesota USA, 1998; p.218.
22. Leslie, J.F. and Summerell . The Fusarium Laboratory Manual. Blackwell Publishing, Ames, IA. 2006; p .388.
23. Cheng, T., Xu, C. Lei, L. Li,. C. Zhang,Y and Zhou, S .Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Molecular Ecology Resources 2016, 16(1), 138-149.
24. White, T.J., T. Bruns, S.J. Lee, W.T and Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 1990, 18(1), 315-322.
25. BIazier, S.R.and Conway, K. E. Characterization of Rhizoctonia solani Isolates Associated With patch diseases on Turfgrass . Pros.Okla. Acad. Sci 2004, 84, 41-51.
26. Dewan, M.M. Identify and frequency of occurrence of fungi in the root of Wheat and ryegrass and their effect on take – and host growth. Ph.D. thesis . Univ . west Australia. 1989;pp .210-240.
27. Mckinney , H.H. Biological control of nematode pests by natural enemies . Ann . Rev. Pytopathol 1923,18 , 415-440.
28. Ramusi, T.M., Aveling, T.A Van der Waals, J.E. and Labuschagne, N. Evaluation of mefenoxam and fludioxonil for control of Rhizoctonia solani, Pythium ultimum and Fusarium solani on cowpea. South African Journal of Plant and Soil 2017, 34(1), 27-33.
29. Gonzalez, M. Pujol, M. Metraux, JP. Gonzalez-Garcia, V. Bolton, MD. Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Molecular plant pathology 2011, 12(3), 209-216.
30. Nzungize, J., Gepts, P. Buruchara, R. Male, A. Ragama, P. Busogoro, J.P. and Baudoin, J.P. Introgression of Pythium root rot resistance gene into Rwandan susceptible common bean cultivars. African Journal of Plant Science . 2011, 5(3),193-200.
31. Makun, H.A., Dutton, M.F Mwanza, M. and Njobeh, P.B. Mycotoxins profiling of the culture material of Fusarium verticillioides (Sacc.) Nirenberg culture (CABI-IMI 392668) isolated from rice in Niger State, Nigeria. African Journal of Biotechnology 2011, 10(56), 12031-12038.
32. Ponukumati, S.V. Elliott, M.L. and Des Jardin, E.A. Comparison of secreted in xylem (SIX) genes in two Fusarium wilt pathogens of ornamental palms. Plant Pathology 2019, 68(9), 1663-1681.
33. Summerell, B.A. Resolving Fusarium: current status of the genus. Annual Review of Phytopathology 2019, 57, 323-339.
34. Arif, M. ; Chawla, S. Zaidi, N. W. Rayar, J. K. Variar, M. and Singh, U. S. Development of specific primers for genus Fusarium and F. solani using rDNA subunit and transcription elongation factor (TEF1) gene. African Journal of Biotechnology 2012, 11(2), 444- 447.
35. Lee, Y.M., Choi, Y.K. and Min, B.R. Molecular characterization of Fusarium solani and its formae speciales based on sequences analysis of the internal transcribed spacer(ITS)region of ribosomal DNA. Mycobiology 2000, 28(2), 82-88.
36. Assuncao, P., Nascimento, L. D. Ferreira, M. F. Oliveira, F. J. Michereff, S. J. and Lima, G. SA.. Reaction of faba bean genotypes to Rhizoctonia solani and resistance stability Iraildes . Horticultura Brasileira 2011, 29, 492-497.
37. Azliza, I. N., Hafizi, R. Nurhazrati, M. and Salleh, B. Production of major mycotoxins by Fusarium species isolated from wild grasses in peninsular Malaysia. Sains Malays 2014, 43, 89-94.
38. Li, S., Hartman, G. L. Lee, B. S. and Widholm,J.W. Identification of a stress-induced protein in stem exudates of soybean seedlings root-infected with Fusarium solani f. sp. glycines. Plant Physiology and Biochemistry 2000, 38(10), 803-809.
39. Toghueo, R.M.K. Bioprospecting endophytic fungi from Fusarium genus as sources of bioactive metabolites. Mycology 2020, 11(1), 1-21.
Received: May 15, 2023/ Accepted: June 10, 2023 / Published: June 15, 2023
Citation: Awad A J, Farhan T A. Isolation and diagnosis of some associated fungi with cowpea root rot disease and testing its pathogenicity. Revis Bionatura 2023;8 (2) 91. http://dx.doi.org/10.21931/RB/2023.08.02.91



















