Vol 7 No 2 2022- 61

1 Fundación PROINPA, Av. Meneces Km. 4, El Paso, Cochabamba, Bolivia.
2 School of Biological Sciences and Engineering, Yachay Tech University, San Miguel de Urcuquí, Imbabura, Ecuador.
3 Independent consultant, Cochabamba, Bolivia.
4 Faculty of Agricultural and Livestock Sciences, Universidad Mayor de San Simón, Cochabamba, Bolivia.
*Corresponding author: jcastillo@yachaytech.edu.ec
Available from: http://dx.doi.org/10.21931/RB/2022.07.02.61
ABSTRACT
Quinoa (Chenopodium quinoa) has grown since ancestral times in the Andean mountains and Altiplano, which are the center of origin of this pseudo-cereal. The interaction of Quinoa with native microorganisms may have contributed to the success of this plant in very adverse climatic and soil conditions. This study addressed the microbial diversity associated with Quinoa plants growing in traditional lands. We employed a cultivable-dependent approach to characterize the communities and identify bacterial strains with potential application in agriculture. We identified bacterial isolates belonging to phyla Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria. The genera Bacillus and Rhizobium/Agrobacterium were the predominant groups in the Quinoa bacterial communities, while various Trichoderma species were also found in the fungi group. The plant growth-promoting ability of selected bacterial strains was assessed by culturing them on media and the in planta test. We used different assays to test the capabilities of the isolates for nitrogen fixation, phosphorus solubilization, and production of the phytohormone indole-3-acetic acid. We inoculated Quinoa seeds with some Bacillus strains and then evaluated plant growth and grain production. Plants inoculated with bacterial strains usually show increased growth parameters and grain yield. Altogether, this work reveals that Quinoa harbors many diverse cultivable bacteria and fungi, which could be used as biological amendments to promote plant growth in a chemical-free way. Avoiding chemical fertilizers helps reduce environmental pollution and maintains the organic character of Quinoa production. International Quinoa markets highly appreciate the organic quality of Quinoa.
Keywords. Plant growth-promoting bacteria, Microbial diversity, Rhizosphere bacteria, Andean Altiplano, Trichoderma
INTRODUCTION
Earth is profusely covered by a great diversity of habitats where the communities of plants and microorganisms live and strive. A particular and extreme ecosystem is the Altiplano, a flat plateau located between 3500 and 4500 m above sea level in the mountain ranges of the Andes. Due to its high altitude, the environmental conditions of Altiplano are severe with hypoxic air and an extreme variety of temperatures and solar radiation1. Its soil is acidic, with little organic matter, shallow and high salinity, especially at Southern Altiplano2. In these conditions, Quinoa (Chenopodium quinoa), a pseudo-cereal native to Andean highlands, grows and produces highly nutritious grains. Human local populations since ancient times domesticated this plant species, which remained genetically unimproved as landraces. Quinoa is highly adapted to the harsh environmental conditions of the Andean Altiplano, including drought, soil salinity, frequent frosts, low soil organic matter, and elevated UV radiation3. Although intensively, Quinoa is still cultivated on the same Altiplano lands that the ancestors used to crop4. Quinoa has lately become a favorite food despite its ancestral status because it does not contain gluten; it is generally organic and highly nutritious (averaging 14.8% protein with an exceptional balance between oil, protein, and starch) 5. This is why Quinoa, the most economically attractive crop in this area, urges Andean farmers to intensify their production beyond sustainability6.
The ability of Quinoa to grow in the rough climatic and edaphic conditions of the Altiplano is due in part to the coexistence with several different microorganisms that interact with it in a mutually beneficial way. Although the information on the microorganisms associated with Quinoa remains scarce, we can assume that some of them are specific to Quinoa and well adapted to this ecosystem7. Therefore, the study of microorganisms associated with Quinoa could give clues about their role in plant nutrition and in diminishing the damaging effects of drought, high soil salinity, and other difficult climatic conditions.
Similarly, the knowledge of the diversity of microorganisms associated with Quinoa opens the possibility of using them for soil restoration, increasing fertility, and pest control. This motivates active research to develop bio-inoculants based on living microorganisms to be used as biofertilizers depending on the microbial species with which they have been made8. The bio inoculants are also essential to reduce the harmful effects of chemical fertilizers and pesticides on the environment, producing food without chemical contaminants at a lower cost, thus helping the economy of small farmers that are common in the Altiplano area9, 7. They could also help the highland soils in the retention of microbial populations that could contribute to delaying degradation processes, allowing farmers to produce organic crops (mainly Quinoa, but also beans, potatoes, and other crops). This is important since organic Quinoa is highly appreciated in international markets10.
The present work assesses the microbial diversity associated with Quinoa plants grown in the Altiplano in the traditional land where Quinoa has been cultivated since ancient time. Because we wish to use the studied microorganisms to improve Quinoa cultivation and its soil, we focused on the cultivable fraction of the microorganisms associated with this species. In this way, we also evaluated the impact of the selected microorganism to be used as biofertilizers for Quinoa. We inoculated Quinoa plants with microorganisms and measured growth parameters, observing a significant increase in the crop’s agronomic parameters, including grain yield.
MATERIALS AND METHODS
Location and sampling.
We selected different land plots with active quinoa production in two provinces of Bolivia, namely Oruro and Potosi, located in the Southern Altiplano of the Andes. Ten plots were considered from communities around the areas of Salinas de Garci Mendoza (19° 38′ 6″ S, 67° 40′ 31″ W, 3,743 m.a.s.l.), Challapata (18° 54′ 2″ S, 66° 46′ 10″ W, 3,718 m.a.s.l.), Pampa Aullagas (19° 11′ 31″ S, 67° 3′ 9″ W, 3,725 m.a.s.l.) and Quillacas (19° 14′ 19″ S, 66° 56′ 21″ W, 3,767 m.a.s.l.) in Oruro province. Similarly, from the province of Potosí 10 plots were selected in the communities of Chacala (20° 33′ 0″ S, 66° 43′ 59″ W, 3,843 m.a.s.l.), and Llica (19° 51′ 4″ S, 68° 14′ 56″ W, 3,672 m.a.s.l.). These communities are located in the southern Altiplano with a semi-arid to the arid climate and temperate summers, cool winters, and predominant dry conditions11. Soils are sandy to sandy-loam, with low water and nutrient retention capacity and very unstable due to the great susceptibility to wind and water erosion; therefore, they tend to desertification. This area has been traditional for quinoa production12.
Microorganism isolation and identification.
Five quinoa plants were collected from each production plot described above. Bacteria and fungi were isolated from different organs of quinoa plants and soil surrounding roots at Fundación Proinpa laboratories, Cochabamba, Bolivia. Bacteria from leaves and stems were isolated by washing plant organs with sterile NaCl 0.85% saline solution and plating samples on a general TSA (tryptic soy agar) medium. To isolate microorganisms from the rhizosphere, we removed the roots and shook the soil closely retained by the roots in a sterile saline solution. Then, bacterial suspension was plated on a semisolid TSA medium. All bacterial cultures were incubated at 28°C for 72 hours13. We thoroughly washed root pieces for endophytic microorganisms and superficially sterilized them by immersing them in a 70% ethanol solution and then in a sodium hypochlorite solution (1.2% v/v). Later, the roots were thoroughly rinsed with sterile saline solution. The roots were macerated in a sterile mortar with saline solution and serial dilutions of 10-1 to 10-8 were deposited on TSA medium and incubated at 28°C for 72h14, 15.
Isolated bacterial colonies were used for species identification. We extracted and purified genomic DNA from bacteria following a standard protocol16. We amplified an internal fragment of the gene encoding the 16S rRNA using primers 27F and 1488R17, 18 and a DNA polymerase with error-correcting activity (Phusion, Finnzymes, Finland). Each PCR product was purified with Qiaquick columns (Qiagen, Valencia, CA) before sending them to the University of Chicago Sequencing Center, Chicago, IL, the USA, for sequencing using Sanger technology. The sequences obtained were edited with the BioEdit program19 and then collated with the databases using the BLAST program20. All 16S rRNA gene sequences obtained in this study were submitted to GenBank under accession numbers MW394536 – MW394612.
Isolated fungi mycelia were used for genomic DNA extraction using the protocol of Melo and collaborators21. DNA fragments corresponding to the genes encoding 18S rRNA, 5.8S rRNA, and the intergenic region (ITS1 and ITS2) were amplified using the SR6RA/T and LR1 primers following the instructions of White and collaborators22, and Kullnig-Gradinger and collaborators23. To further discriminate between possible Trichoderma species, we amplified an internal fragment of the tef1 gene that encodes the elongation factor 1 alpha using the EF1-728F and TEF1LLErev primers24. The amplified fragments were purified and sequenced, as explained above. The sequences were analyzed using the TrichOKey program, which allows sequences of different marker genes to be used to determine the Trichoderma species to which an isolate corresponds25.
Phylogenetic Analysis
The sequences generated during microorganism identification were trimmed with Bioedit software19. Multiple sequence alignment was performed with the MAFFT program through its web interface at https://mafft.cbrc.jp/alignment/server/. The phylogenetic tree was inferred by the Mega X program26 using the Maximum Likelihood method and General Time Reversible model. Subtree-Pruning-Regrafting was used as the heuristic method, and 1,000 resamples of bootstrap were tested. The tree was drawn with Dendroscope 3.5.927. Accession numbers of the reference strains used in the phylogeny are given in Table 2.
Table 2. Accession numbers of the reference strains used in the phylogenetic reconstruction.
Plant growth-promoting (PGP) characterization.
We characterized several plant growth-promoting traits as ecosystem services that quinoa-associated microorganisms may provide to agriculture and their potential use as bio inoculants. For nitrogen fixation, we have grown the bacteria on Burk’s N-free culture medium28 and incubated the bacteria for 5 to 15 days at 28°C. All plates with bacterial growth were considered bacterial isolates capable of fixing nitrogen. For phosphorous solubilization, bacteria were plated on NBRIP medium with tricalcium phosphate as a source of P29. Bacterial strains that showed a transparent halo around the colony were recorded as positive. For testing indole-3-acetic acid (IAA) production, we followed the protocol according to Gordon and Weber30 with slight modifications. Briefly, bacterial colonies were grown in TSB (soy trypticase broth) supplemented with 5μM of L-tryptophan and incubated for 7 days at 28°C (bacteria), under agitation (100 rpm). After gentle centrifugation to discard cells and debris, the supernatant was transferred to 96-well microtiter plates and supplemented with Salkowski solution (15 ml 0.5 M FeCl3, 500 ml distilled water, and 300 ml of concentrated H2S0431) that allows for detecting indole derivatives. We used two well-identified Bacillus species as a positive control. The mixture was kept in the dark for 15 min, and all wells turning color to pink or fuchsia were reported as positive.
Plant growth parameters and grain yield were tested in quinoa plants inoculated with the beneficial bacteria isolated from Quinoa. We used the landrace of Quinoa called Horizontes (breed variety with short plants, high yield, and large, white grains). Three seeds were sown in a sterile substrate (1:1:1:2 manure, sand, lama, and rice husk) in pots under greenhouse conditions (16±2°C, 90% RH, and 11:13 light-dark relationship, which simulates the winter photoperiod in the Altiplano). Concomitantly with planting, the seeds were inoculated with 500 µl of the bacterial isolate concentrate (by pouring bacteria suspension in sterile distilled water at 1×108 spores/ml). The plants were regularly watered and remained in the greenhouse until full production of grains (about 6 months).
We measured the following plant growth parameters and yield: Root length (cm), root volume (ml), root, foliage, and panicle weight (g), plant height (cm), panicle length and diameter (cm), and grain yield (kg/ha). These parameters were measured at the end of the growth cycle except plant height, which was recorded after 90 days. All of the measured parameters were subjected to analysis of variance using the SAS software32. The sources of variation considered were amendment treatments with different beneficial bacteria: Bacillus sp. 13b (Subtilis group), Paenibacillus sp. 15c, Bacillus sp. 17c (Subtilis group), Bacillus sp. 3b (Subtilis group), Bacillus sp. 6b (Subtilis group), and Bacillus sp. 12c (Cereus group) (all of these strains were collected in Challapata, Oruro province, except 13b, which was isolated from Llica). We used sterile distilled water with no bacteria as a negative control. The chosen experimental design is a completely random block model with seven factors (bacteria strains and control) and 6 repetitions of one pot with 3 plants each. Differences between treatments were estimated using one-way ANOVA, and the average values for each parameter were compared with each other using the Tukey test at p<0.05.
RESULTS
Many bacterial isolates (1000-2000) were obtained from each Altiplano community (see Location and Sampling in the Materials and Methods section). Analysis of the bacterial communities of Quinoa resulted in the identification of 77 isolates (Table 1) corresponding to 4 phyla and 17 genera (Table 1). Figure 1A shows the relative abundance of phyla. The most frequently isolated group was Proteobacteria which accounted for 46.8%. This group includes representatives of three classes (Alphaproteobacteria, Gammaproteobacteria and Betaproteobacteria) and the principal genera are Rhizobium/Agrobacterium. As the second most dominant bacterial phylum, Firmicutes amounts for 43% of the total bacteria sampled. The relative contribution of the other two phyla, Bacteroidetes and Actinobacteria, is minor. At the genus level, the molecular identification was sufficient to determine the taxonomic affiliation of most isolates; except for Rhizobium and Agrobacterium whose taxonomic classification is still unclear33. The most common bacterial genera were Bacillus (35%), followed by Rhizobium/Agrobacterium (32.5%). Several genera, such as Phyllobacterium, Ensifer, and others, only contained one isolate (Fig. 1B). Unfortunately, identifying bacterial isolates to species level was not possible for all samples due to the remarkable similarity of 16S rRNA sequences between phylogenetically close species. This difficulty applies mainly to the isolates belonging to the genus Bacillus; however, it is also seen in other groups. Despite the low discriminatory power of 16S rRNA, we were able to identify many Bacillus isolates belonging to well-known internal phylogenetic groups of the genus Bacillus according to the recent phylogenetic reconstruction of this genus34, 35. Therefore, the Quinoa isolates in the Cereus clade (Table 1) most probably correspond to B. cereus, B. thuringiensis, B. toyonensis or other species of this genus. The Quinoa isolates grouped in the Subtilis clade presumably are B. subtilis, B. amyloliquefaciens, B. velezensis, B. vallismortis, B. tequilensis, or B. licheniformis. For the case of Pumilus clade, its bacterial members isolated from Quinoa may correspond to B. safensis, B. altitudinis or B. pumilus. The taxonomic affiliations of novel bacteria isolated in this study are confirmed by a phylogenetic study depicted in Fig. 2. We used 16S rRNA gene sequences of representative strains of each bacterial species/genus to highlight the correct taxonomic assignation of Quinoa isolates in the phylogenetic tree.

Figure 1. The relative abundance of phyla (A) and genera (B) is associated with Quinoa plants grown in the Bolivian Altiplano.

Figure 2. 16S rRNA gene sequences phylogram (maximum likelihood) of bacteria associated with Quinoa plants sampled from original lands in the Bolivian Altiplano. Sequences from representative genomes are in bold. Node labels are bootstrap values. The scale bar represents the number of nucleotide substitutions per site.

Table 1. Cultivable microorganisms associated with Quinoa and their taxonomic information.
For the case of fungi, we concentrated our efforts on isolating and identifying species belonging to Trichoderma for their agricultural potential to use them as plant growth promoters and biocontrol agents. Based on the analyses of ribosomal regions and tef1 gene sequences, fungi isolates were identified as different species of Trichoderma (Table 1). Although we have used different marker genes to differentiate Trichoderma species, the discrimination was not enough to differentiate isolates from T. asperellum and T. koningiopsis. We did not infer the phylogeny of the fungi associated with Quinoa because the analysis was restricted to very few species of the same genus.
Plant growth-promoting characterization.
Although we did not analyze all the isolates sampled, most of them were examined for their properties as plant growth promoters. Figure 3 shows the relative proportion of quinoa bacterial strains with important functional traits for agriculture. The most frequent traits were the production of IAA and P solubilization, which were observed for 53.3% and 30.8% of the analyzed strains, respectively. Although most bacterial isolates showed a single PGP trait, some strains of Bacillus (Subtilis and Pumilus groups) and Paenibacillus showed different traits simultaneously.
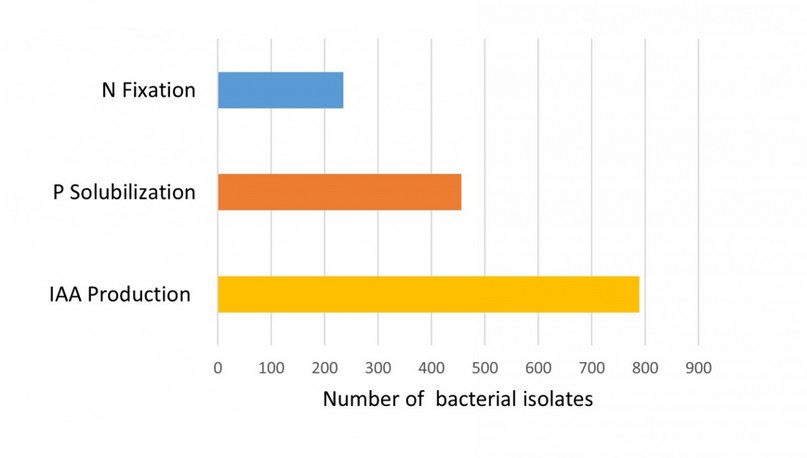
Figure 3. Plant growth-promoting (PGP) traits in bacteria associated with Quinoa.
We selected six bacterial strains from Bacillaceae family that showed the highest response in tests of PGP traits to analyze their plant growth promotion and grain production in quinoa plants grown under greenhouse conditions. The six strains show single or multiple PGP traits as analyzed by in vitro tests (see Materials and Methods, Table 3). We inoculated quinoa seeds with the bacterial strains and measured different parameters until the end of the quinoa production cycle. Figure 4 compares root, foliage, and panicle growth in control plants versus plants treated with Bacillus sp. 12b, Bacillus sp. 13b, Paenibacillus sp. 15c, Bacillus sp. 17c, Bacillus sp. 3b, Bacillus sp. 6b. Only the treatments with Bacillus sp. 12b, Paenibacillus sp. 15c, Bacillus sp. 17c, and Bacillus sp. 6b showed a statistically significant increase in plant growth parameters versus the control treatments. In contrast, both Bacillus sp. 13b and Bacillus sp. 3b showed no significant effect. Figure 5 compares plant height, panicle length, and diameter in these same plants. Here the performance of some treatments is inferior compared to the control; however, Bacillus sp. 6b shows consistency in being superior to the others in the measured parameters.

Figure 4. Effect of amendment applications of beneficial bacteria on Quinoa plants to test plant growth and grain yield. Specifically, it is shown variation in foliage, root and panicle weight. Bars that do not share similar letters denote statistical significance at p=0.05 with one-way ANOVA and Tukey’s test. The bars at the top of each histogram represent a statistical error.

Figure 5. Effect of amendment applications of beneficial bacteria on quinoa plants to test plant growth and grain yield. Specifically, it is shown variation in plant height, panicle length and diameter. Bars that do not share similar letters denote statistical significance at p=0.05 with one-way ANOVA and Tukey’s test. The bars at the top of each histogram represent a statistical error.

Table 3. Main PGP traits shown by selected bacterial isolates were analyzed for their ability to promote the growth of Quinoa plants in the greenhouse.
In contrast, there was no statistical difference for root length between any of the treatments and the control (data not shown), and a similar situation is observable in root volume, with treatments behaving equal or worse than the control (Fig. 6). Figure 7 shows the results of the bacterial amendments in the quinoa grain yield regarding control plants. Again, Bacillus sp. 12b and Bacillus sp. 6b showed a statistically significant higher yield of grain versus the control treatment.
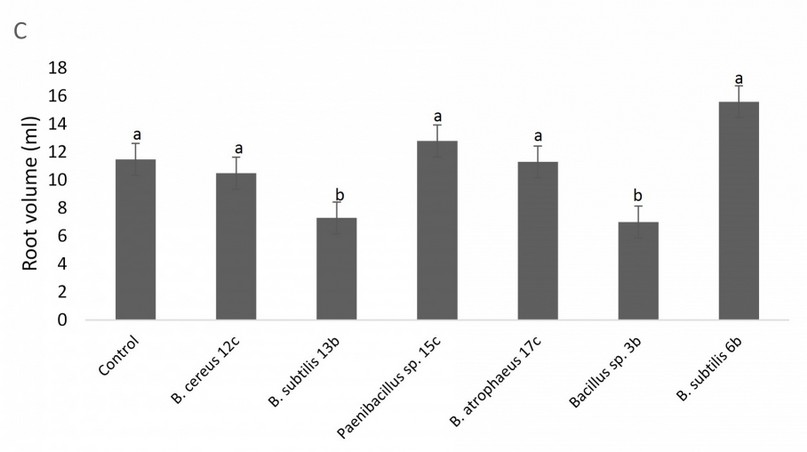
Figure 6. Effect of amendment applications of beneficial bacteria on Quinoa plants to test plant growth and grain yield. Specifically, it is shown variation in root volume. Bars that do not share similar letters denote statistical significance at p=0.05 with one-way ANOVA and Tukey’s test. The bars at the top of each histogram represent a statistical error.

Figure 7. Effect of amendment applications of beneficial bacteria on Quinoa plants to test plant growth and grain yield. Specifically, it is shown variation in grain yield. Bars that do not share similar letters denote statistical significance at p=0.05 with one-way ANOVA and Tukey’s test. The bars at the top of each histogram represent a statistical error.
DISCUSSION
This study adopted a culture-dependent strategy to analyze microbial diversity associated with Quinoa. We focused on the cultivable fraction because we intend to use the sampled microorganisms to enrich the natural microbiota and increase plant growth and grain production in Quinoa plots. Likewise, microorganisms are promising for the appropriate management of dryland ecosystems (such as the Andean Altiplano) and for reducing soil degradation36, 37. Although the diversity of bacteria was not investigated by any culture-independent technique, the culture-dependent method remains a valuable tool for understanding the diversity of microorganisms interacting with Quinoa in the singular ecosystem of the Altiplano. In this work, we extended previous investigations carried out by our group7, including new data to highlight the abundant microbial diversity associated with Quinoa and some results about the plant-promoting capacity of Quinoa microorganisms. We profusely isolated bacterial samples. Although we did not identify all the microorganisms collected, those identified indicate the diversity associated with Quinoa. The diversity includes four phyla, Firmicutes (mainly Bacillus), Actinobacteria, Bacteroidetes, and Proteobacteria. In all analyzed samples, the genera Bacillus and Rhizobium/Agrobacterium were the predominant groups in the Quinoa bacterial communities. The low discriminatory power of 16S rRNA gene sequences is insufficient to identify Bacillus species, which is increasingly common in many environmental and medical samples38, 39. However, we confirmed the affiliation of samples to groups inside the Bacillus genus according to the current phylogeny described by Patel and Gupta34and Secaira-Morocho and collaborators35. Notably, Bacillus was also found as a Quinoa endophyte by Pitzschke40, in Quinoa soils by Morales-Belpaire and collaborators41, and in the rhizosphere of Quinoa roots by Liceta42.
The dominance of Bacillus in the Quinoa microbial communities may be due to the natural ability to form endospores that allows it to survive in very inhospitable conditions such as those of the Altiplano43. The presence of Bacillus associated with Quinoa has greater significance since this bacterial genus actively intervenes in germination and the seedling establishment, reducing at the same time the stress damage by activating different enzymes and elicitors40, 44. In general, Bacillus species show different PGP activities that may include nitrogen fixation, phosphate solubilization, siderophore release, and the production of plant growth regulators such as auxins (IAA), cytokinins, gibberellins, ethylene, and abscisic acid. Likewise, other indirect mechanisms may work concomitantly, including the production of inhibitory substances (antibiotics, hydrolytic enzymes, etc.), which act against phytopathogens, increasing the natural resistance45. Our work found that the Bacillus strains showed some of these mechanisms (i.e., N fixation, P solubilization, and IAA production).
Regarding other rhizobacteria isolated from Quinoa roots, they are well-known dwellers of the roots microbiome, helping the plant face abiotic stresses. These bacteria exhibit tolerance to multiple physiological or environmental conditions. For example, Psychrobacter sp. Stenotrophomonas maltophilia and Paenibacillus xylanexedens are psychrotolerant species46, 47, 48 that could contribute reducing plant cold damage due to the freezing temperatures of the Altiplano reaches during winters49. On the other hand, some Variovorax paradoxus, Pseudomonas fluorescens, Rhizobium, and Pantoea are recognized as halotolerant plant-growth-promoting rhizobacteria50, 51, 52 that may contribute to increasing salt tolerance in Quinoa plants. Since some natural soils where Quinoa grows are saline6, the presence of these strains is not surprising. Lastly, bacterial strains of Microbacterium sp., Pedobacter sp. and spore formers (Bacillus, Paenibacillus) are UV tolerant53, 54, 55 a trait that is incredibly important to tackle with the high solar radiation and low humidity content that is typical in the highlands.
Our study concerning plant growth promoted by bacteria under greenhouse conditions found that some PGP activities significantly stimulated plant growth and grain production. In particular, Bacillus sp. 6b (Subtilis clade) showed the highest promotion activity in terms of the growth parameters compared with the control, including the foliage, root, panicle weight, and plant height panicle length and diameter, root volume, and grain yield. Interestingly, Bacillus sp. 6b showed N fixation capability but no appreciable IAA production or P solubilization; therefore, additional PGP mechanisms of this strain remain to be discovered. Many members of the Bacillus subtilis group (i.e. B. subtilis, B. amyloliquefaciens, B. licheniformis, and others) are well-known plant-growth-promoting rhizobacteria and may activate their plant host defense responses56. These bacteria may also act as biocontrol agents against pathogens because of their ability to secrete antibiotics, hydrolytic enzymes, and lipopeptides, thus suppressing plant diseases57, 58. Another strain of Bacillus that showed good performance in the plant growth promotion and grain yield in our assays was Bacillus sp. 12c (Cereus group). Although the Cereus group comprises well-known food-borne pathogens that can cause infectious gastrointestinal and respiratory diseases in humans59, they are also known for their ability to promote plant growth60. For example, B. cereus may be involved in promoting plant growth and may also participate in conferring tolerance to the plant host to reduce stress in saline environments. Hassan and collaborators61 found that B. cereus works to ameliorate the deleterious effects of salinity. The soil where Quinoa plants thrive is often saline, especially in the southern Altiplano. In this sense, the protective role that B. cereus may offer to Quinoa plants could be essential for life under saline stressful conditions. Paenibacillus sp. 15c, another bacterial species with good performance on plant growth promotion, showed P solubilization and IAA production in our tests. This is not surprising since Paenibacillus species are recognized residents of plant rhizosphere that promote plant growth by different mechanisms that include the production of IAA and other auxin phytohormones62.
Given the results of this work, microorganisms with PGP activities have enormous potential to be used in sustainable agriculture to promote plant growth and control pathogens63. Although we have not carried out tests to find out the capacity of the isolated strains to control pathogens, it is very likely that Quinoa-associated bacteria also show this trait. The use of environmentally friendly bacteria to substitute chemical fertilizers and avoid losing productive soil in semi-desert lands is desirable and promising. This is even more important considering that most of the Quinoa produced in the Altiplano is organic and traded in European markets64, 65; therefore, this crop should remain free of agrochemicals.
CONCLUSIONS
In this work, we characterized the diversity of microorganisms associated with Quinoa using a cultivable-dependent method and tested the capacity of the isolated microorganisms to promote quinoa growth and increase grain production. The abundant diversity of bacteria in Quinoa could help to understand the critical role that microorganisms play in quinoa growth and grain production. We probed in this work that the addition of quinoa-derived bacteria to seeds during the sown process shows beneficial effects on plant growth and grain yield. These results are promising and can give rise to further research towards the biotechnological application of beneficial microorganisms as biofertilizers, contributing by then to keeping the organic character of quinoa production that is highly required in international markets.
Acknowledgments
This research was carried out by scientists from the Fundación PROINPA with economic resources from the “Regional Fund for Agricultural Technology” (FONTAGRO) grant number 7075 and the “Andean Consortium”, supported by the General Directorate for International Cooperation of the Netherlands.
Declaration of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
1. Garreaud, R.D., 2009. The Andes climate and weather. Advances in Geosciences 22:3–11. https://doi.org/10.5194/adgeo-22-3-2009
2. Senra, E.O., Schaefer, C. E., Corrêa, G. R., Gjorup, D. F., Reis, J. S., & Francelino, M. R., 2019. Holocene pedogenesis along a chronotoposequence of soils from the Altiplano to the Cordillera Real, Bolivian Andes. CATENA, 178, 141–153. doi:10.1016/j.catena.2019.03.012 10.1016/j.catena.2019.03.012
3. Garcia, M., Raes, D., Jacobsen, S.-E., 2003. Evapotranspiration analysis and irrigation requirements of Quinoa (Chenopodium quinoa) in the Bolivian highlands. Agricultural Water Management 60:119–134. https://doi.org/10.1016/S0378-3774(02)00162-2
4. Cruz, P., Winkel, T., Ledru, M.-P., Bernard, C., Egan, N., Swingedouw, D., Joffre, R., 2017. Rain-fed agriculture thrived despite climate degradation in the pre-Hispanic arid Andes. Science Advances 3:e1701740. https://doi.org/10.1126/sciadv.1701740
5. Wright, K.H., Pike, O.A., Fairbanks, D.J., Huber, C.S., 2002. Composition of Atriplex hortensis, Sweet and Bitter Chenopodium quinoa Seeds. Journal of Food Science 67:1383–1385. https://doi.org/10.1111/j.1365-2621.2002.tb10294.x
6. Jacobsen, S.-E., 2011. The Situation for Quinoa and its Production in Southern Bolivia: From Economic Success to Environmental Disaster: Quinoa Production in Southern Bolivia. Journal of Agronomy and Crop Science 197:390–399. https://doi.org/10.1111/j.1439-037X.2011.00475.x
7. Ortuño N, Claros M, Gutiérrez C, Angulo M, Castillo JA. 2014. Bacteria associated with the cultivation of quinoa in the Bolivian Altiplano and their biotechnological potential. Revista de Agricultura (Bolivia) 53:53-61.
8. Pathak, D.V., Kumar, M., 2016. Microbial Inoculants as Biofertilizers and Biopesticides. In: Microbial Inoculants in Sustainable Agricultural Productivity, eds Singh, D.P., Singh, H.B., Prabha, R. New Delhi: Springer. https://doi.org/10.1007/978-81-322-2647-5_11
9. Arora, N.K. 2018. Agricultural sustainability and food security. Environmental Sustainability 1:217–9. https://doi.org/10.1007/s42398-018-00032-2
10. Furche C, Salcedo S, Krivonos E, Rabczuk P, Jara B, Fernández D, 2013. International quinoa trade. In: State of the art report on quinoa around the world in 2013, eds Bazile, D., Bertero, H.D., Nieto, C. Rome: FAO/CIRAD.
11. Monasterio, M.; Andressen, L. R.; Terceros, L. F. 2007. Regímenes climáticos del altiplano sur de Bolivia: una región afectada por la desertificación. Revista Geográfica Venezolana 48:11-32.
12. Orsag V, Castro E, Patzi L, León ML, Pacosaca O, Mamani F. 2011. Evaluación de fertilidad de los suelos en la zona intersalar: Producción sostenible de quinua. 1st ed. La Paz: Fundación PIEB.
13. Hagedorn, C., Gould, W.D., Bardinelli, T.R., 1989. Rhizobacteria of Cotton and their Repression of Seedling Disease Pathogens. Applied and Environmental Microbiology 55:2793–2797. https://doi.org/10.1128/AEM.55.11.2793-2797.1989
14. McInroy, J.A., Kloepper, J.W., 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant and Soil 173:337–342. https://doi.org/10.1007/BF00011472
15. Magallón, P.; Dion, P. 2009. Importancia de los microorganismos promotores de crecimiento vegetal para los pequeños productores de Bolivia. Practical-theoretical course on agricultural microbiology. Bolivia: Fundación Proinpa.
16. Wilson K. 1994. Preparation of genomic DNA from bacteria. In Current protocols in molecular biology, eds. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. New York: John Wiley and Sons.
17. Lane, D. J. 1991. 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics, eds. Stackebrandt E. and Goodfellow M. New York: John Wiley and Sons.
18. Herrera-Cervera, J.A., Caballero-Mellado, J., Laguerre, G., Tichy, H.-V., Requena, N., Amarger, N., Martinez-Romero, E., Olivares, J., Sanjuan, J., 1999. At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiology Ecology 30:87–97. https://doi.org/10.1111/j.1574-6941.1999.tb00638.x
19. Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/N.T. Nucleic Acids Symposium Series 41:95–98.
20. Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. 1990. Basic Local Alignment Search Tool. Journal of Molecular Biology 215:403-410
21. Melo, S.C.O., Pungartnik, C., Cascardo, J.C.M., Brendel, M., 2006. Rapid and efficient protocol for DNA extraction and molecular identification of the basidiomycete Crinipellis perniciosa. Genetics and Molecular Research 5:851-5.
22. White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications, eds. Innis MA, Gelfand DH, Sninsky JJ, White TJ., San Diego: Academic Press.
23. Kullnig-Gradinger, C.M., Szakacs, G., Kubicek, C.P., 2002. Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycological Research 106:757–767. https://doi.org/10.1017/S0953756202006172
24. Nagy, V., Seidl, V., Szakacs, G., Komoń-Zelazowska, M., Kubicek, C.P., Druzhinina, I.S., 2007. Application of DNA Bar Codes for Screening of Industrially Important Fungi: the Haplotype of Trichoderma harzianum Sensu Stricto Indicates Superior Chitinase Formation. Applied and Environmental Microbiology 73:7048–7058. https://doi.org/10.1128/AEM.00995-07
25. Druzhinina, I.S., Kopchinskiy, A.G., Komoń, M., Bissett, J., Szakacs, G., Kubicek, C.P., 2005. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genetics and Biology 42:813–828. https://doi.org/10.1016/j.fgb.2005.06.007
26. Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K., 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35:1547–1549. https://doi.org/10.1093/molbev/msy096
27. Huson, D.H. Richter, D.C., Rausch, C. Dezulian, T., Franz, M., Rupp, R. 2007. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460.
28. Park, M., Kim, C., Yang, J., Lee, H., Shin, W., Kim, S., Sa, T., 2005. Isolation and characterization of diazotrophic growth promoting bacteria from rhizosphere of agricultural crops of Korea. Microbiological Research 160:127–133. https://doi.org/10.1016/j.micres.2004.10.003
29. Nautiyal, C.S., 1999. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiology Letters 170:265–270. https://doi.org/10.1111/j.1574-6968.1999.tb13383.x
30. Gordon, S.A., Weber, R.P., 1951. Colorimetric estimation of Indoleacetic Acid. Plant Physiology 26:192‐195, 1951.
31. Meudt, W.J., Gaines, T.P., 1967. Studies on the Oxidation of Indole-3-Acetic Acid by Peroxidase Enzymes. I. Colorimetric Determination of Indole-3-Acetic Acid Oxidation Products. Plant Physiology 42:1395–1399. https://doi.org/10.1104/pp.42.10.1395
32. SAS, 2000. SAS software. SAS Institute Inc., Cary, NC, USA.
33. Flores-Félix, J.D., Menéndez, E., Peix, A., García-Fraile, P., Velázquez, E., 2020. History and current taxonomic status of genus Agrobacterium. Systematic and Applied Microbiology 43, 126046. https://doi.org/10.1016/j.syapm.2019.126046
34. Patel, S., Gupta, R.S. 2020. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. International Journal of Systematic and Evolutionary Microbiology 70:406–38.
35. Secaira-Morocho, H., Castillo, J.A., Driks, A., 2020. Diversity and evolutionary dynamics of spore-coat proteins in spore-forming species of Bacillales. Microbial Genomics 6:1 https://doi.org/10.1099/mgen.0.000451
36. Turan, M., Esitken, A., Sahin, F., 2012. Plant Growth Promoting Rhizobacteria as Alleviators for Soil Degradation. In: Bacteria in Agrobiology: Stress Management, ed. Maheshwari, DK. Berlin Heidelberg: Springer. https://doi.org/10.1007/978-3-662-45795-5_3
37. Pointing, S.B., Belnap, J. 2012. Microbial colonization and controls in dryland systems. Nature Reviews Microbiology 10:551–62. https://doi.org/10.1038/nrmicro2831
38. Srinivasan, R., Karaoz, U., Volegova, M., MacKichan, J., Kato-Maeda, M., Miller, S., Nadarajan, R., Brodie, E.L., Lynch, S.V., 2015. Use of 16S rRNA Gene for Identification of a Broad Range of Clinically Relevant Bacterial Pathogens. PLoS ONE 10:e0117617. https://doi.org/10.1371/journal.pone.0117617
39. Johnson, J.S., Spakowicz, D.J., Hong, B.-Y., Petersen, L.M., Demkowicz, P., Chen, L., Leopold, S.R., Hanson, B.M., Agresta, H.O., Gerstein, M., Sodergren, E., Weinstock, G.M., 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nature Communications 10:5029. https://doi.org/10.1038/s41467-019-13036-1
40. Pitzschke, A., 2016. Developmental Peculiarities and Seed-Borne Endophytes in Quinoa: Omnipresent, Robust Bacilli Contribute to Plant Fitness. Frontiers in Microbiology 7. https://doi.org/10.3389/fmicb.2016.00002
41. Morales-Belpaire, I., Miranda-Torrez, G., Mendez-Pinaya, V., Morales, C., Elliot-Portal, E., Justo-Chipana, I. 2011. Producción de biofertilizantes para quinua con bacterias fijadoras de nitrógeno. Embajada Real de Dinamarca. La Paz: Fudación PIEB.
42. Liceta M. 2015. Aislamiento y caracterización de Pseudomonas y Bacillus provenientes de la rizósfera de diferentes variedades de quinua (Chenopodium quinoa Willd.) y su uso como potenciales promotoras del crecimiento vegetal. Degree thesis. Universidad Nacional Agraria La Molina, Peru.
43. Nicholson, W.L., Munakata, N., Horneck, G., Melosh, H.J., Setlow, P., 2000. Resistance of Bacillus Endospores to Extreme Terrestrial and Extraterrestrial Environments. Microbiology and Molecular Biology Reviews 64:548–572. https://doi.org/10.1128/MMBR.64.3.548-572.2000
44. Pitzschke, A., 2018. Molecular dynamics in germinating, endophyte-colonized quinoa seeds. Plant and Soil 422:135–154. https://doi.org/10.1007/s11104-017-3184-2
45. Sansinenea, E., 2019. Bacillus spp.: As Plant Growth-Promoting Bacteria. In: Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms, eds Singh, H.B., Keswani, C., Reddy, M.S., Sansinenea, E., García-Estrada, C. Singapore: Springer. https://doi.org/10.1007/978-981-13-5862-3_11
46. Bozal, N., Montes, M.J., Tudela, E., Guinea, J., 2003. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp. nov. and Psychrobacter fozii sp. nov. International Journal of Systematic and Evolutionary Microbiology 53:1093–1100. https://doi.org/10.1099/ijs.0.02457-0
47. Vazquez, S., Ruberto, L., Mac Cormack, W., 2005. Properties of extracellular proteases from three psychrotolerant Stenotrophomonas maltophilia isolated from Antarctic soil. Polar Biology 28, 319–325. https://doi.org/10.1007/s00300-004-0673-6
48. Nelson, D.M., Glawe, A.J., Labeda, D.P., Cann, I.K.O., Mackie, R.I., 2009. Paenibacillus tundrae sp. nov. and Paenibacillus xylanexedens sp. nov., psychrotolerant, xylan-degrading bacteria from Alaskan tundra. International Journal of Systematic and Evolutionary Microbiology 59:1708–1714. https://doi.org/10.1099/ijs.0.004572-0
49. López-Moreno, J.I., Morán-Tejeda, E., Vicente-Serrano, S.M., Bazo, J., Azorin-Molina, C., Revuelto, J., Sánchez-Lorenzo, A., Navarro-Serrano, F., Aguilar, E., Chura, O., 2016. Recent temperature variability and change in the Altiplano of Bolivia and Peru: Recent Temperature Variability and Change in the Altiplano. International Journal of Climatology 36:1773–1796. https://doi.org/10.1002/joc.4459
50. Ahmad, M., Zahir, Z.A., Nazli, F., Akram, F., Arshad, M., Khalid, M., 2013. Effectiveness of halo-tolerant, auxin producing Pseudomonas and Rhizobium strains to improve osmotic stress tolerance in mung bean (Vigna radiata L.). Brazilian Journal of Microbiology 44:1341–1348. https://doi.org/10.1590/S1517-83822013000400045
51. Gond, S.K., Torres, M.S., Bergen, M.S., Helsel, Z., White, J.F., 2015. Induction of salt tolerance and up-regulation of aquaporin genes in tropical corn by rhizobacterium Pantoea agglomerans. Letters in Applied Microbiology 60:392–399. https://doi.org/10.1111/lam.12385
52. Zhou, N., Zhao, S., Tian, C.-Y., 2017. Effect of halotolerant rhizobacteria isolated from halophytes on the growth of sugar beet (Beta vulgaris L.) under salt stress. FEMS Microbiology Letters 364:fnx091 https://doi.org/10.1093/femsle/fnx091
53. Arrage, A.A., Phelps, T.J., Benoit, R.E., White, D.C., 1993. Survival of subsurface microorganisms exposed to UV radiation and hydrogen peroxide. Applied and Environmental Microbiology 59:3545–3550. https://doi.org/10.1128/AEM.59.11.3545-3550.1993
54. Correa-Llantén, D.N., Amenábar, M.J., Blamey, J.M., 2012. Antioxidant capacity of novel pigments from an Antarctic bacterium. Journal of Microbiology 50:374–379. https://doi.org/10.1007/s12275-012-2029-1
55. Reis-Mansur, M.C.P.P., Cardoso-Rurr, J.S., Silva, J.V.M.A., de Souza, G.R., Cardoso, V. da S., Mansoldo, F.R.P., Pinheiro, Y., Schultz, J., Lopez Balottin, L.B., da Silva, A.J.R., Lage, C., dos Santos, E.P., Rosado, A.S., Vermelho, A.B., 2019. Carotenoids from UV-resistant Antarctic Microbacterium sp. LEMMJ01. Scientific Reports 9:9554. https://doi.org/10.1038/s41598-019-45840-6
56. Hashem, A., Tabassum, B., Fathi Abd_Allah, E., 2019. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi Journal of Biological Sciences 26:1291–1297. https://doi.org/10.1016/j.sjbs.2019.05.004
57. Cazorla, F.M., Romero, D., Pérez-García, A., Lugtenberg, B.J.J., Vicente, A. de, Bloemberg, G., 2007. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity: Characterization of antagonistic Bacillus. Journal of Applied Microbiology 103:1950–1959. https://doi.org/10.1111/j.1365-2672.2007.03433.x
58. Mnif, I., Ghribi, D. 2015. Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications: Lipopeptides Biosurfactants and their Applications. Biopolymers 104:129–47. https://doi.org/10.1002/bip.22630
59. Kotiranta, A., Lounatmaa, K., Haapasalo, M., 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes and Infection 2:189–198. https://doi.org/10.1016/S1286-4579(00)00269-0
60. Tilak, K. V. B. R.; Srinivasa Reddy B. 2006. Bacillus cereus and B. circulans – novel inoculants for crops. Current Science 90:642-644.
61. Hassan T, Naz I, Hussain M. 2018. Bacillus cereus: A competent plant growth promoting bacterium of saline sodic field. Pakistan Journal of Botany 50:1029-37.
62. Grady, E.N., MacDonald, J., Liu, L., Richman, A., Yuan, Z.-C., 2016. Current knowledge and perspectives of Paenibacillus: a review. Microbial Cell Factories 15:203. https://doi.org/10.1186/s12934-016-0603-7
63. Verma, M., Mishra, J., Arora, N.K., 2019. Plant Growth-Promoting Rhizobacteria: Diversity and Applications. In: Environmental Biotechnology: For Sustainable Future, eds. Sobti, R.C., Arora, N.K., Kothari, R. Singapore: Springer. https://doi.org/10.1007/978-981-10-7284-0_6
64. Laguna, P., Cáceres, Z., Carimentrand, A., 2006. Del altiplano sur boliviano hasta el mercado global: Coordinación y estructuras de gobernancia en la cadena de valor de la quinua orgánica y del comercio justo. Agroalimentaria 22:65-76,
65. Jaldín Quintanilla, R., 2010. Producción de quinua en Oruro y Potosí. Estados de investigación temática PIEB. Programa de Investigación Estratégica en Bolivia, La Paz: Fundación PIEB.
Received: 20 January 2022 / Accepted: 9 May 2022 / Published:15 May 2022
Citation. . Castillo J A, Conde G , Claros M, Ortuño N. Diversity of cultivable microorganisms associated with Quinoa (Chenopodium quinoa) and their potential for plant growth-promotion. Revis Bionatura 2022;7(2) 61. http://dx.doi.org/10.21931/RB/2022.07.02.61






















