Vol 7 No 2 2022- 27

Zahraa A. Sharba1 and Halah Farazdaq Rafeeq 2,*
1 Department of Applied Sciences / University of Technology/Iraq; Zahraa.A.Sharba@uotechnology.edu.iq
2 Department of Applied Sciences / University of Technology/Iraq; halahfr@gmail.com
* Correspondence: halahfr@gmail.com; Tel.: +9647722299212
Available from: http://dx.doi.org/10.21931/RB/2022.07.02.27
ABSTRACT
In community-acquired urinary tract infections, Klebsiella pneumoniae is considered one of the most common etiological agents. Multidrug resistance and virulence are common in Klebsiella pneumoniae populations. In this study, fifty Klebsiella pneumoniae isolates were isolated from urine samples and identified using a vitek 2 compact device. The Kirby–Bauer disk diffusion technique used the antibiotic susceptibility test. According to the findings, approximately [n = 46 (92%)] of Klebsiella pneumoniae isolates are multidrug-resistant (MDR). To detect the production of Extended Spectrum β-lactamase (ESBL) enzymes, the Modified Double Disc Synergy Test (MDDST) was used. The results show that approximately [n=45 (90%)] of the isolates produce ESBLs. The most common ESBL genes (TEM, SHV, and CTX-M) were investigated in isolates. The results show that the SHV gene had the highest prevalence among ESBL genes [n = 34 (68%)], followed by the CTX-M gene [n = 33 (66%)]. while none of the isolates possessed the TEM gene. The virulence factor type 3 fimbriae (MrKD) gene and biofilm (BssS) gene were revealed. The results found that the isolates contain the MrKD gene at [n = 41 (82%)]. At the same time, the results found that the isolates contained the BssS gene at [n =36 (72%)]. The prevalence of Virulence genes within ESBL-producing Klebsiella pneumoniae isolates shows that only [n = 3 (6%)] of isolates that are non-ESBL producers carry one or both virulence genes, while [n=41 (82%)] of ESBL-producing isolates contain one or both virulence genes. The prevalence of ESBL-producing Klebsiella pneumoniae in community patients was high in this research. There may also be a correlation between ESBL production and some virulence factors.
Keywords. Klebsiella pneumoniae; Antibiotic Resistance; Virulence Gene; ESBL; Urinary Tract Infection; CTX-M.
INTRODUCTION
Urinary tract infections (UTIs) have severe health and financial consequences for society. These infections are considered the most prevalent type of bacterial infection, and they can strike at any moment during a person’s life. People in hospitals and the general public are susceptible to urinary tract infections. CAUTIs (community-associated urinary tract infections) are frequently identified in individuals with risk factors such as age, prior UTI history, sexual activity, and diabetes mellitus. Antibiotics are the most common therapy for bacterial UTIs1. After E.coli, K. pneumoniae is the most medically significant species in the Enterobacteriaceae family2. One of the frequent pathogens linked to both community and hospital-acquired urinary tract infections is K. pneumoniae3. Because bacterial etiology is common in UTIs, broad-spectrum antibiotics are frequently used, resulting in a rise of resistant uropathogens1. Antibiotic resistance among K. pneumoniae strains is a major public health concern and a significant financial burden for UTI sufferers. Antibiotic resistance of these isolates is acquired by various processes, including the creation of extended spectrum-lactamases (ESBLs). The formation of extended-spectrum beta-lactamase, an enzyme that attacks the beta-lactam ring in medicines and renders them useless, causes ESBL resistance4. Several ESBL families have been identified; however, the prominent families such as SHV, TEM, and CTX-M account for the bulk of ESBLs5. The rapidly increasing resistance of ESBL producers to multiple antibiotic families is a significant issue that limits the treatment options available to ESBL producers6. In addition, lipopolysaccharide (LPS), capsular polysaccharide, adhesions, and siderophores are among the virulence components found in K. pneumoniae that contribute to its pathogenicity7. Adhesion is an essential phase in the infection that must be strong enough to overcome the host’s defensive mechanisms. Fimbriae, also known as pili, are proteinaceous structures that extend from the bacterial cell surface to a distance of 100 nm to several microns and are composed of adhesions that are thought to aid bacterial adhesion8. There are two kinds of fimbriae or pili in K. pneumoniae, types 1 and 39. Type 3 fimbriae have a helix-like structure that gives them spring-like flexibility and stretch-ability. This fimbrial type is encoded by a chromosomally borne gene cluster that has previously been demonstrated to consist of five genes in various strains of K. pneumoniae. The mrkABCDF gene cluster encodes the primary structural component (mrkA) and the fimbrial adhesin (mrkD), whereas the genes mrkB and mrkC encode the chaperone and usher proteins, respectively10. Type 3 fimbriae have been shown to mediate adhesion to various structures in human kidney and lung tissue and epithelial cells from human urine sediments and endothelial and bladder epithelial cell lines in vitro investigations11. K. pneumoniae biofilm development on biotic and abiotic inert surfaces is commonly linked to Type 3 fimbriae12. Infections caused by K. pneumoniae producing biofilms are more resistant to therapy than other infections. Biofilms serve as shields for bacterial populations, allowing them to evade host defenses. The bacteria are also protected from severe circumstances such as pH changes, forces of shear, and nutritional deficiencies13. As a result, the current study was designed to look into the distribution of extended-spectrum β-lactamase enzyme genes (TEM, SHV, CTX-M), virulence genes (type 3 fimbriae MrKD gene, biofilm BssS gene), and the relationship between them in K. pneumoniae isolates from urine samples.
MATERIALS AND METHODS
Bacterial Isolation and Identification
From different laboratories in Baghdad, (50) isolates of K. pneumoniae were collected from (133) bacterial isolates from urine samples from the period (2020/07/1) to (2020/09/25). For their identification, K. pneumoniae isolates were diagnosed by the vitek 2compact (bioMérieux/France) device.
Antibiotic Susceptibility Test
The Kirby–Bauer disk diffusion technique was used to perform Antibiotic susceptibility testing for all isolates14. Antibiotics listed below were utilized: Ampicillin (10 𝜇g), Piperacillin (100 𝜇g), Amoxicillin-Clavulanate (20/10 𝜇g), Ceftazidime/Avibactam (30/20 𝜇g), Cefotaxime (30 𝜇g), Ceftriaxone (30 𝜇g), Ceftazidime (30 𝜇g), Cefuroxime (30 𝜇g), Cefazolin (30 𝜇g), Loracarbef (30 𝜇g), Cepodoxime (10 𝜇g), Cefixime (5 𝜇g), Aztreonam (30 𝜇g), Imipenem (10 𝜇g), Meropenem (10 𝜇g), Gentamicin (10 𝜇g), Tobramycin (10 𝜇g), Amikacin (30 𝜇g), Doxycycline (30 𝜇g), Tetracycline (10 𝜇g), Ciprofloxacin (5 𝜇g), Levofloxacin (5 𝜇g), Nalidixic acid (30 𝜇g), Trimethoprim-Sulfamethoxazole (1.25/23.75 𝜇g), Chloramphenicol (30 𝜇g), Nitrofurantoin (300 𝜇g).
Modified Double Disc Synergy Test (MDDST)
Detect ESBL enzymes production by using the Modified Double Disc Synergy Test through the following steps15: After uniformly spreading the inoculum onto sterile Mueller–Hinton agar (biolab/Hungary), the disc which contained amoxicillin-Clavulanate (20/10µg) was placed in the center of the plate and the discs of third-generation cephalosporin [cefotaxime (30µg), ceftriaxone (30µg), and cefpodoxime (10µg)], and fourth-generation cephalosporin [cefepime (30µg)], were placed 15mm and 20 mm apart respectively, center to center to that the amoxicillin-Clavulanate disc. After being Incubated for 18–24 h at 37 °C, any increase in the inhibition zone toward the amoxicillin-Clavulanate disc was considered positive for ESBL production.
Detection of Antibiotic Resistance and Virulence Factor Genes
DNA extraction
For the isolation and purification of DNA from K. pneumoniae isolates, DNA was extracted using G-spinTM Total DNA Extraction Kit (Intron biotechnology/Korea) according to the manufacturer’s instructions. Using Nanovue plus™ spectrophotometer (GE Healthcare/UK), estimated concentration and purity.
Polymerase Chain reaction of Antibiotic Resistance and Virulence Genes
The PCR method detects genes encoding virulence factors (type 3 fimbriae MrKD, biofilm BssS) and extended-spectrum -lactamase genes (TEM, SHV, CTX-M). Primers used in this study were purchased from (Alpha DNA/Canada) in lyophilized form, as shown in Table 1. The PCR amplification reaction mixture components (20µl) used to detect each gene contain the following: DNA sample, primers, nuclease-free water, and 5x FIREPol® Master Mix (Solis Bio Dyne/Europe) 16. After preparing the reaction volume in the PCR tube, the mixture was spun down via (Centrifuge/Vortex for PCR plates) and then PCR tubes were placed in the PCR thermal cycler (Bio-Rad/USA) to amplify the target DNA for (TEM, SHV, CTX-M, BssS, MrKD primers) using the following program17 as shown in Table 2.
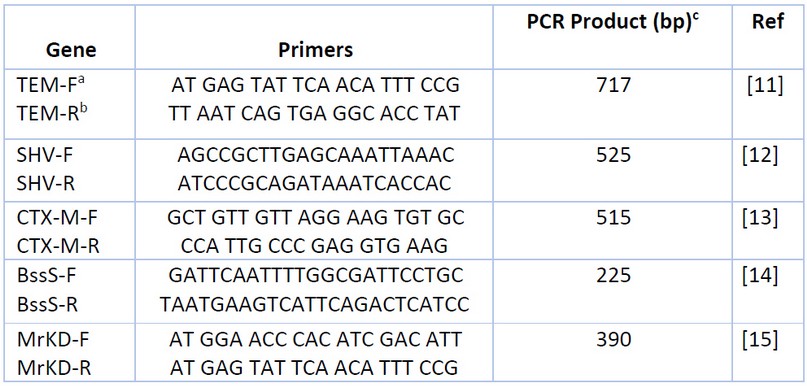
Table 1. Primers Used for Detection Genes.

Table 2. PCR Thermocycler Conditions for Each Gene.
RESULTS
In the current study, 50 K. pneumoniae isolates were collected from urine samples of community-acquired urinary tract infection patients. All samples were identified by vitek 2compact device (bioMérieux/France).
Antibiotic Susceptibility Test
The antibiotic susceptibility test was done by the Kirby–Bauer disk diffusion method, then interpreted the results were using criteria published by the Clinical and Laboratory Standard Institute18. As seen in Table 3. The results showed that about [n=46(92%)] of K. pneumoniae isolates were multidrug resistance (MDR).

Table 3. Antibiotic Susceptibility of 50 K. pneumoniae Isolates.
The Modified Double Disc Synergy Test (MDDST) was used to detect ESBL production. As a result, any increase in the inhibition zone of cefeime, cefpodoxime, ceftriaxone, and cefotaxime toward the amoxicillin clavulanate disc was interpreted as a sign of ESBL generation. The results showed that approximately [n=45 (90%)] of the isolates produced ESBLs, as shown in Figure 1.

Figure 1. Modified Double Disc Synergy Test (MDDST) (A) shows synergism of cefeime, cefpodoxime, ceftriaxone and cefotaxime with amoxicillin-Clavulanate, which indicates ESBL production, while in (B) showed no synergism with amoxicillin-Clavulanate, which indicates none of ESBL production.
Detection of Antibiotic Resistance and Virulence Factors Genes
The ability of K. pneumoniae isolates to produce ESBLs (TEM, SHV, and CTX-M) genes and the capacity of isolates to generate Virulence factor type 3 fimbriae (MrKD) genes and biofilm (BssS) genes are determined using PCR, which uses sets of primers that amplify the genes. The results showed that the SHV gene was the most prevalent among ESBL genes [n=34(68%)], then the CTX-M gene [n=33(66%)]. While none of the isolates possessed the TEM gene, as showed in Figure 2.

Figure 2. Agarose gel electrophoresis of PCR products of (A) TEM gene, (B) SHV gene and (C) CTX-M gene in K. pneumoniae isolates visualized under UV after staining by RedSafeTM nucleic acid staining solution for 2% agarose gel at 100 volts, 70 amps for 60 min. L: ladder DNA (100pb), S: Sample.
The results found that the isolates contain MrKD gene at [n=41 (82%)] as shown in the figure A (3). It also, found that [n=36 (72%)] of isolates contain the BssS gene as shown in the figure B (3).

Figure 3. Agarose gel electrophoresis of PCR products of (A) MrKD gene and (B) BssS gene in K. pneumoniae isolates visualized under UV after staining by RedSafeTM nucleic acid staining solution for 2% agarose gel at 100 volts, 70 amps for 60 min. L: ladder DNA (100pb), S: Sample.
The prevalence Virulence genes within ESBL-producing K. pneumoniae isolates show that only [n=3 (6%)] of isolates which are non-ESBL producing carry one or both virulence genes, while [n=41 (82%)] of ESBL-producing isolates contain one or both virulence genes as seen in Table 4.

Table 4. Distribution Virulence genes within ESBLs and non-ESBLs producing K. pneumoniae isolates.
DISCUSSION
In both community and hospital settings, K. pneumoniae is a leading cause of serious infections such as urinary tract infection, pneumonia, skin and soft tissue infection, intra-abdominal infection, bloodstream infection, meningitis, and pyogenic liver abscess. In humans, antimicrobials have long been used to treat K. pneumoniae infections19. The emergence of multidrug-resistant (MDR) K. pneumoniae strains worldwide is a major source of worry20. In this study, the results showed that about [n=46(92%)] of K. pneumoniae isolates were multidrug resistance (MDR). MDR is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories21. Different rates of MDR were reported in many studies, such as the study done by Mohamed et al., which reported that K. pneumoniae had the highest level of resistance to conventional antibiotics, with 86.66% of isolates being MDR22. Also, a study done by Pishtiwan and Khadija concluded that most of the K. pneumoniae isolates were multidrug-resistant (MDR). Where 75% of K. pneumoniae isolates showed the MDR phenotype23. MDR obstructs disease control by increasing the risk of resistant microorganisms spreading, lowering treatment efficacy and, as a result, causing patients to be infected for more extended periods of time24. A variety of virulence factors contribute to K. pneumoniae’s pathogenicity 25. As well as the ability to rapidly develop antibiotic resistance 26. K. pneumoniae is, in fact, a significant ESBL host. The development of ESBLs has significantly increased bacterial resistance to beta-lactams in human infections, causing significant morbidity and death. 27. Extended-spectrum beta-lactamases (ESBLs) are enzymes that can give resistance towards β-lactam antibiotics such penicilins, scephalosporins, and aztreonam. This is achieved by hydrolyzing the antibiotics, and beta-lactamase inhibitors like clavulanic acid block these enzymes28.
This study used Modified Double Disc Synergy Test (MDDST) to detect ESBL production. The results showed that approximately [n=45 (90%)] of the isolates produced ESBLs. These results were close to those obtained by Shakib et al., who showed that [n=62 (88.6%)] of K. pneumoniae isolates were ESBL-producing29. Pishtiwan and Khadija et al.’s research revealed that [n=17(85%)] of K. pneumoniae isolates were positive for ESBL production23. The prevalence of ESBL-producing clinical isolates is related to risk factors such as current antibiotic use and hospitalization 29.
The ability of K. pneumoniae isolates to produce ESBLs (TEM, SHV, and CTX-M) genes showed that the SHV gene was the most prevalent among ESBL genes [n=34(68%)], then the CTX-M gene [n=33(66%)]. At the same time, none of the isolates possessed the TEM gene. The study done by Malekjamshidi et al. showed that the most prevalent β-lactamase gene was SHV, followed by TEM and CTX-M 30. Also, Pishtiwan and Khadija et al. reveal that the TEM gene has the highest prevalence in K. pneumoniae isolates at 64.7%, followed by the CTX-M gene at 41.1% and the SHV gene at 35.2% 23. In addition, a study by Mbelle et al. found that all isolates contained the CTX-M gene, while (97%) contained TEM, and (83%) harbored SHV genes 31. There are significant geographic differences in the prevalence of resistant bacteria and their genes. Low-and middle-income countries generally have higher endemic antimicrobial resistance than high-income countries, mainly driven by antibiotic overuse in humans and animals 32. International travel can facilitate the transfer of resistant bacteria and their genes from their endemic regions to other locations 33.
CONCLUSION
The capacity of K. pneumoniae isolates to generate Virulence factor type 3 fimbriae (MrKD) genes and biofilm (BssS), found that the isolates contain MrKD gene at [n=41 (82%)]. While the results found that [n=36 (72%)] of isolates contain the BssS gene. The prevalence Virulence genes within ESBL-producing K. pneumoniae isolates show that only [n=3 (6%)] of isolates that are non-ESBL producing carry one or both virulence genes, while [n=41 (82%)] of ESBL-producing isolates contain one or both virulence genes. This is consistent with the study done by Gharrah et al. that suggests a correlation between ESBL production and some virulence factors34. In conclusion, the prevalence of ESBL-producing K. pneumoniae in community patients was high in this research. It’s also possible that ESBL production and virulence factors are linked. Antibacterial resistance is a developing problem in clinical practice. As a result, improved clinical awareness and laboratory testing are required to decrease treatment failure and prevent the spread of ESBL-producing K. pneumoniae.
Funding: This research received no external funding.
REFERENCES
1. Foxman, B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infectious Disease Clinics 2014, 28, 1–13. https://doi.org/10.1016/j.idc.2013.09.003.
2. Lai, C.C.; Lee, K.; Xiao, Y.; Ahmad, N.; Veeraraghavan, B.; Thamlikitkul, V.; et al. High burden of antimicrobial drug resistance in Asia. Journal of Global Antimicrobial Resistance2014. 2, 141–147. https://doi.org/10.1016/j.jgar.2014.02.007.
3. Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiology and Molecular Biology Reviews 2016, 80, 629–661. https://doi.org/10.1128/MMBR.00078-15.
4. Mohsen, S.M.; Hamzah, H.A.; Imad Al-Deen, M.M.; Baharudin, R. Antimicrobial susceptibility of Klebsiella pneumoniae and Escherichia coli with extended-spectrum β-lactamase associated genes in Hospital Tengku Ampuan Afzan, Kuantan, Pahang. The Malaysian journal of medical sciences: MJMS 2016, 23, 14–20.
5. Naseer, U.; Sundsfjord, A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microbial drug resistance 2011, 17, 83–97. http://doi.org/10.1089/mdr.2010.0132.
6. Pitout, J. D.; Laupland, K. B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. The Lancet infectious diseases 2008, 8, 159-166. https://doi.org/10.1016/S1473-3099(08)70041-0.
7. Lery, L.M.; Frangeul, L.; Tomas, A.; Passet, V.; Almeida, A.S.; Bialek-Davenet, S.; et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC biology 2014, 12, 1, 1-15.
8. Murphy, C.N.; Clegg, S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future microbiology 2012, 7, 991-1002. https://doi.org/10.2217/fmb.12.74.
9. Nakkash, A.F.; Al-Husseiny, K.R. Study Some Virulence Factors of Klebsiella pneumoniae Isolated from Clinical Sources. J. Thi-Qar Univ 2008, 3, 45-50.
10. Stahlhut, S.G.; Chattopadhyay, S.; Kisiela, D.I.; Hvidtfeldt, K.; Clegg, S.; Struve, C.; et al. Structural and Population Characterization of MrkD, the Adhesive Subunit of Type 3 Fimbriae. Journal of bacteriology 2013, 195, 5602-5613. https://doi.org/10.1128/JB.00753-13.
11. Schroll, C.; Barken, K.B.; Krogfelt, K.A.; Struve, C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC microbiology 2010, 10, 1-10.
12. Alwan, A.H.; Abass, S.M. The effects of U.V light on mrkA, mrkD genes in local isolates of Klebsiella pneumoniae. Al-Mustansiriyah Journal of Science 2016, 27, 1-6. http://dx.doi.org/10.23851/mjs.v27i4.23.
13. Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrobial Resistance and Infection Control 2019, 8, 1-10. https://doi.org/10.1186/s13756-019-0533-3.
14. Vandepitte, J.; Verhaegen, J.; Engbaek, K.; Piot, P.; Rohner, P.; Heuck, C.C. Basic Laboratory Procedures in Clinical Bacteriology, 2nd ed.; World Health Organization: Geneva, Switzerland, 2003, pp. 103-121.
15. Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: a Clinical Update. Clinical microbiology reviews 2020, 18, 657-686. https://doi.org/10.1128/CMR.18.4.657-686.2005.
16. Sharba, M.M.; Al-janabi, A.A. SEQUENCE AND STRUCTURE ANALYSIS OF HLA-G IN BREAST CANCER PATIENT USING BIOINFORMATICS TOOLS AND TECHNIQUES. Connect journals 2021, 1, 617-622.
17. Abed, S.A.; Al-Khafaji, H.M.; Design of Primer and Probe to Detect SNP Rs 1892901 in Fosl-1Gene in Different Types of Cancer in Iraqi Population. Systematic Re-views in Pharmacy 2021, 12, 296-300.
18. Weinstein, M.P.; Krin, T.J.; Limbago, B.; LewisII, J.S.; Bobenchick, A.M.; Moters,A.J.; et al. Performance standards for antimicrobial susceptibility testing, 30th ed.; Clinical and Laboratory Standards Institute : Pennsylvania, USA, 2020, 1-15.
19. Vading, M.; Nauclér, P.; Kalin, M.; Giske, C.G. Invasive infection caused by Klebsiella pneumoniae is a disease affecting patients with high comorbidity and associated with high long-term mortality. PloS one 2018, 13, e0195258. https://doi.org/10.1371/journal.pone.0195258.
20. Struve, C.; Forestier, C.; Krogfelt, K.A. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 2003, 149, 167-176. https://doi.org/10.1099/mic.0.25833-0.
21. Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection 2012, 18, 268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
22. Mohamed, S.H.; Khalil, M.S.; Mohamed, M.S.M.; Mabrouk, M.I. Prevalence of antibiotic resistance and biofilm formation in Klebsiella pneumoniae carrying fimbrial genes in Egypt. Res J Pharm Technol 2020, 13, 3051-3058.
23. Pishtiwan, A.H.; Khadija, K.M. Prevalence of blaTEM, blaSHV, and blaCTX-M Genes among ESBL-Producing Klebsiella pneumoniae and Escherichia coli Isolated from Thalassemia Patients in Erbil, Iraq. Mediterranean journal of hematology and infectious diseases 2019, 11, e2019041. Doi: 10.4084/MJHID.2019.041.
24. Carson, C.F.; Hammer, K.A.; Riley, T.V. Broth micro-dilution method for determining the susceptibility of Escherichia coli and Staphylococcus aureus to the essential oil of Melaleuca alternifolia (tea tree oil). Microbios 1995, 82,181-185.
25. Victor, L.Y.; Hansen, D.S.; Ko, W.C.; Sagnimeni, A.; Klugman, K.P.; Von Gottberg, A.; et al. Virulence Characteristics of Klebsiella and Clinical Manifestations of K. pneumoniae Bloodstream Infections. Emerging infectious diseases 2007, 13, 986-993. Doi: 10.3201/eid1307.070187.
26. Kumar, V.; Sun, P.; Vamathevan, J.; Li, Y.; Ingraham, K.; Palmer, L.; et al. Comparative Genomics of Klebsiella pneumoniae Strains with Different Antibiotic Resistance Profiles. Antimicrobial Agents and Chemotherapy 2011, 55, 4267-4276. Doi: https://doi.org/10.1128/AAC.00052-11.
27. Atmani, S.M.; Messai, Y.; Alouache, S.; Fernández, R.; Estepa, V.; Torres, C.; Bakour, R. Virulence characteristics and genetic background of ESBL-producing Klebsiella pneumoniae isolates from wastewater. Fresenius Environmental Bulletin 2015, 24,103-112.
28. Ben-Ami, R.; Rodríguez-Baño, J.; Arslan, H.; Pitout, J.D.; Quentin, C.; Calbo, E. S.; Carmeli, Y. A Multinational Survey of Risk Factors for Infection with Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Non-hospitalized Patients. Clinical Infectious Diseases 2009, 49, 682-690. https://doi.org/10.1086/604713.
29. Shakib, P.; Kalani, M.T.; Ramazanzadeh, R.; Ahmadi, A.; Rouhi, S. Molecular detection of virulence genes in Klebsiella Pneumoniae clinical isolates from Kurdistan Province, Iran. Biomedical Research and Therapy 2018, 5, 2581-2589. DOI: 10.15419/bmrat.v5i8.467.
30. Malekjamshidi, M.R.; Zandi, H.; Eftekhar, F. Prevalence of Extended-Spectrum β-lactamase and Integron Gene Carriage in Multidrug-Resistant Klebsiella Species Isolated from Outpatients in Yazd, Iran. Iranian journal of medical sciences 2020, 45, 23-31. Doi: 10.30476/IJMS.2019.45334.
31. Mbelle, N.M.; Feldman, C.; Sekyere, J.O.; Maningi, N.E.; Modipane, L.; Essack, S.Y. Pathogenomics and evolutionary epidemiology of multidrug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Scientific reports 2020, 10, 1-17. | https://doi.org/10.1038/s41598-020-58012-8.
32. Klein, E.Y.; Tseng, K.K.; Pant, S.; Laxminarayan, R. Tracking global trends in the effectiveness of antibiotic therapy using the Drug Resistance Index. BMJ global health 2019, 4, e001315. http://dx.doi.org/10.1136/bmjgh-2018-001315.
33. Frost, I.; Van Boeckel, T.P.; Pires, J.; Craig, J.; Laxminarayan, R. Global geographic trends in antimicrobial resistance: the role of international travel. Journal of travel medicine 2019, 26, taz036. https://doi.org/10.1093/jtm/taz036.
34. Gharrah, M.M.; El-Mahdy, A.M.; Barwa, R.F. Association between virulence factors and extended spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdisciplinary Perspectives on Infectious Diseases 2017, 2017, 1-14. https://doi.org/10.1155/2017/7279830.
Received: 5 December 2021 / Accepted: 29 January 2022 / Published:15 May 2022
Citation: Sharba Z, Farazdaq Rafeeq H. The Incidence of Extended Spectrum β-Lactamase Enzymes and Their Connection to Virulence Genes in Community-Acquired Urinary Tract Infection. Revis Bionatura 2022;7(2) 27. http://dx.doi.org/10.21931/RB/2022.07.02.27



















