Vol 7 No 1 2022- 11

Major Genetic Determinants of Extended-Spectrum Β-Lactamase (ESBL), Carbapenemase, Fosfomycin and Colistin Resistance in Escherichia Coli from Intensive Care Units
1 Dept. of Med. and Mol. Biotech., College of Biotechnology, Al-Nahrain University, Baghdad, Iraq 1
2 DNA forensic Center for Research and Training, Al-Nahrain University, Baghdad, Iraq 2; Marwa.gen13@yahoo.com
3 Post Graduate, Dept. of Quality Control, Grain Board of Iraq, Ministry of Trade, Baghdad, Iraq 2; omarnatheer77@yahoo.com
* Correspondence: e-mail; alshammariahmed.a.m@gmail.com
Available from: http://dx.doi.org/10.21931/RB/2022.07.01.11
ABSTRACT
Escherichia coli (E. coli) strains placed in predominant nosocomial bacteria in intensive care units (ICUs), resulting in severe drug-resistant infections. Non-susceptibility to β-lactams and last-line drugs such as Fosfomycin and Colistin cause limited availability of infections eradication. The objective of this study included the determination of genes encoding extended-spectrum β-lactamase (ESBL), Carbapenemase, Colistin, and Fosfomycin resistance in clinical isolates of E. coli in ICUs. A total of 200 E. coli isolates were identified from ICU settings. The CTXM-1, SHV, IMP and OXA-48 genes were detected for β-lactamases using the polymerase chain reaction (PCR) technique. The fosA3 and mcr-1 and mcr-2 genes were also detected for resistance against Fosfomycin and Colistin. The CTX-M1, SHV, IMP and OXA-48 genes were detected in 60 (30%), 56 (28%), 28 (14%) and 4 (8%) of isolates. none of the E. coli isolates had the mcr-2 and fosA3 genes. Despite the existence of resistance genes to the third-generation antibiotics and Carbapenemase s, any isolates had genes for resistance to Fosfomycin and Colistin. More studies are needed to follow the resistance genes against last-resort antibiotics.
Keywords. Escherichia coli, beta-lactamase, Fosfomycin, Colistin, ESBLs.
INTRODUCTION
Escherichia coli (E. coli) organism is known as the cause of wound and urinary infections, pneumonia, meningitis, sepsis, etc., so it is unique along with other members of the normal intestinal flora 1-5. This bacterium is facultatively anaerobic, oxidase negative and catalase-positive. E. coli is one of the most common microorganisms in the normal intestinal flora of humans and warm-blooded animals 6-9. It colonizes in the host intestine a few days after birth. Today, the increasing spread of antibiotic-resistant bacteria is one of the problems of human society. Bacteria can be resistant to different types of antibiotics through different mechanisms 10-13. One of the most common antibiotic resistance mechanisms, especially in Gram-negative bacteria, is the production of beta-lactamase enzymes. They are enzymes that cause the beta-lactam antibiotic ring’s cleavage, such as Penicillins and cephalosporins, to inactivate these drugs, thereby protecting the bacteria against the drug.
Extended-spectrum beta-lactamase (ESBL) enzymes are beta-lactamases that hydrolyze Penicillins, first-, second-, and third-generation cephalosporins, as well as aztreonam, but do not affect carbapenems (such as imipenem) and cephamycins (such as cefoxitin) 7,10,11.
Despite the measures that have been taken in developing broad-spectrum antimicrobials, the emergence and spread of bacterial resistance is a concern 1,3,6,10,11. Gram-negative spp is one of the significant barriers to properly treating infectious diseases. Among them, those that produce ESBLs restricted treatment of menacing infections. E. coli is a common species in the production of ESBL enzymes and is a major cause of nosocomial infections 14,15. On the other hand, resistance to carbapenems is a severe issue that is caused by various enzymes. Intensive care units are basic settings where the resistant strains spread. Major ESBL and Carbapenemase genes include CTX-M, SHV, TEM, IMP, VIM and OXA-48, which are spread with various resistance levels 1,3,10,11,14. The aim of our study included detection of genes encoding ESBLs (SHV and CTX-M1), Carbapenemase s (OXA-48 and IMP1), Colistin and Fosfomycin among E. coli from ICU settings.
MATERIALS AND METHODS
Bacterial isolates and patients
450 stool samples were initially collected and cultured onto the blood agar and MacConky media (MErk, Germany). Suspected colonies were subjected to the IMVIC test to identify them. These tests included indole production, motility in the SIM medium, vogues pro square and citrate consumption. 200 E. coli was determined using biochemical tests from ICU settings. Patients included 84 males and 112 females with an age mean of 52.21 years. All the patients had more than 24h of hospitalization.
Phenotypic detection of ESBLs
The production of ESBLs enzymes was confirmed using the synergy and combine tests. The synergy test included a culture of isolate onto the Mueller Hinton agar and placing two disks co-amoxiclav and ceftazidime near each other. Additionally, the combined disk included placement of ceftazidime disk and a blank disk that contained clavulanic acid. Any increase in the inhibition zone exhibited the production of ESBLs.
DNA isolation
The total genomic DNA was extracted using the boiling method. After preparation of bacterial suspension from a single colony of isolates in DDH2O, each tube was boiled for 10min and centrifuged at 10,000rpm for 5min. The supernatant was used as a DNA template. The quantification of the DNA load was evaluated using the 260/280 ratio and Nanodrop device to be >1.8.
Detection of ESBLs and Carbapenemase genes
The ESBL and Carbapenemase genes, including CTX-M, SHV, IMP and OXA-48 were detected using PCR and using specific primers shown in table 1. The PCR conditions included 94°C for 3min, and a 30 cycle of 94°C, annealing for 45s, 72°C for 1min and final extension of 72°C for 10min. The master mix (6µL), template DNA (1µL), each F and R primer (1µL) and DDH2O (5µL) were mixed in the reaction tube.

Table 1. Specific primers for detection of CTX-M, SHV, IMP and OXA-48 genes.
Detection of mcr-1, mcr-2 and fosA3 genes
The PCR assay was employed for the amplification of Fosfomycin (fosA3) and Colistin (mcr-1, mcr-2) resistant genes using specific primer sequences and annealing shown in table 2. 19.

Table2. The primer sequences for the detection of Fosfomycin and Colistin resistance genes.
The thermal cycle for detecting mcr-1 and mcr-2 genes was similar to that of previous, but the annealing temperature included 56°C and 58°C. The PCR products were run onto the 1% gel agarose in 1X Tris Borate EDTA (TBE) and visualized using safe dye.
Data analysis
The prevalence of genes was analyzed by SPSS version 20, and the un-paired T-test and ANOVA options were used for assessment of significance defining <0.05 cut-off value.
RESULTS
Patients and isolates
Of 200 patients, 84 males and 112 females with a mean age of 52.21 years were included. The previous hospitalization was determined in 177 of them (p<0.001). All the patients had more than 24h of hospitalization. Moreover, 189 (p<0.0001) of them had previous beta-lactam administration. The age range of patients and rate of ESBLs production has been depicted in table3. 70 patients were infected with ESBL-producing E. coli.
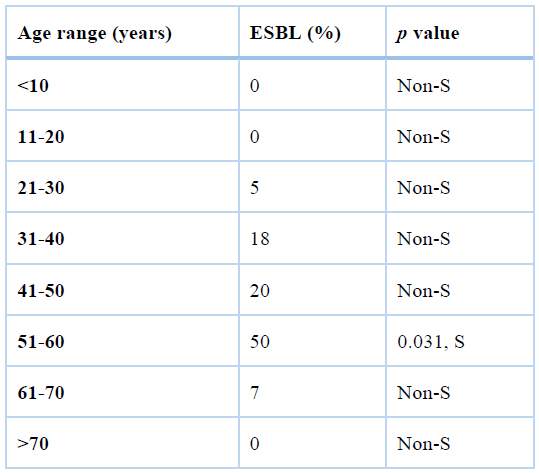
S: significant
Table3. The age range of patients and ESBL production.
As shown in table3, the age range 51-60 years were significantly more infected with ESBL-producing E. coli, but other age groups were not significantly associated with the ESBL production.
ESBL and Carbapenemase genes
The CTX-M1, SHV, IMP and OXA-48 genes were detected in 60 (30%), 56 (28%), 28 (14%) and 4 (8%) of isolates (figures 1-4).

Figure 1. PCR gel electrophoresis of CTX-M1 with 500 bp size.

Figure 2. PCR gel electrophoresis of SHV with 1016 bp size.

Figure 3. PCR gel electrophoresis of IMP with 198 bp size.

Figure 4. PCR gel electrophoresis of OXA-48 gene with 484 bp size.
Notably, three isolates had all the CTX-M1, SHV, IMP, and OXA-48 genes. The patients had ages>60 years and had previous hospitalization and antibiotic consumption.
The association of ESBL genes with previous hospitalization and antibiotic prescription has been depicted in table 4. As shown, there was a significant relationship between the previous hospitalization and antibiotic prescription and the rate of ESBL production (p<0.0001).

Table 4. The association of ESBL genes with previous hospitalization and antibiotic prescription.
PCR detection of mcr-1, mcr-2 and fosA3 genes
None of the E. coli isolates had the mcr-1, mcr-2 and fosA3 genes.
DISCUSSION
Despite the fact that infectious diseases and their treatment have always been considered throughout human history, their changes and mutations have caused failure in their eradication 1,6,10,11. On the other hand, even this has caused new strains, and their prevalence has increased the range of these diseases in new forms. Microbial strains which produce enzymes hydrolyzing broad-spectrum antimicrobials are a concern. Gram-negative species are one of the significant barriers to the definitive treatment of infectious diseases 20-22. E. coli is a common species in the production of ESBL enzymes and is the main cause of nosocomial infections, especially urinary tract infections and sepsis, viscera, liver abscesses, cholangitis and cholecystitis and pancreatic abscesses. Therefore, due to the growing number of these strains in nosocomial infections and ICU centers, determining the pattern of E. coli resistance genes encoding ESBLs and Carbapenemase enzymes in clinical samples was one of the objectives of this study.
In this study, the rate of CTX-M SHV ESBLs included 30% (n=60) and 28% (n=56), respectively. The production of ESBL enzymes is encoded by several genetic factors that produce over 340 different types of beta-lactamase enzymes. These enzymes are classified into different classes based on their genetic identification and similarity. There are different reports of the prevalence of these enzymes in the Enterobacteriaceae family in the world. Studies in other parts of the world have reported a different prevalence. It seems that the emergence and spread of ESBL-bearing bacteria are often due to the widespread use of beta-lactam drugs so that today we see an increasing number of these bacteria in various parts of hospitals such as ICUs. In a study by Tasli et al. in Turkey, the production of ESBL in E. coli strains was equal to 17% 23, and in the study of Villegas in Colombia, it was reported 3.4% 24.
On the other hand, the Zhou study in Shanghai showed that 47.4% of E. coli isolated from patients produced these enzymes 19. In another study by Wu et al. in Taiwanese hospitals, ESBL-producing E. coli was 18.18% as one of the most abundant ESBL-producing isolates 25. In Lebanon, the rate was 28.1%. This rate is much lower in Japan, less than 0.1% in E. coli and 0.3% in Klebsiella spp. Elsewhere in Asia, it has been reported from 4.8% to 28%25,26. Comparison of these results shows that the rate of ESBL in isolated strains from different countries and in one country from one hospital to another varies depending on the infection control system and patient’s treatment method in each hospital. Various risk factors are involved in increasing the number of ESBL-producing bacteria, such as long-term hospital stay, overuse of antibiotics (including third-generation cephalosporins), use of vascular and urinary catheters, and prolonged ICU storage history of surgery and irrational and inadequate use of antimicrobial treatments. This study also observed that prior hospitalization and antibiotic usage are significant risk factors.
We also observed that the IMP and OXA-48 genes were detected in 28 (14%) and 4 (8%) isolates, respectively. Fortunately, none of the isolates had genes encoding Fosfomycin (fosA3) and Colistin (mcr-1 and mcr-2) resistance.
CONCLUSIONS
In this study, a high rate of E. coli isolates from ICU were ESBL-producing by amplifying CTX-M and SHV genes. Fortunately, none of the isolates had genes encoding Fosfomycin (fosA3) and Colistin (mcr-1 and mcr-2) resistance. More studies are needed to follow the resistance genes against last-resort antibiotics.
Informed Consent Statement: Not applicable.
Data Availability Statement: The results of this study will be found under the rules of the journal and related link.
Acknowledgments: To my colleagues working in laboratories inside my college and to those who helped me collect samples and resources.
Conflict of interest: None to declare
REFERENCES
1. Hamadamin HA, Baban ST, Hussen BM, Ali FAJJoKMCV. Distribution of blaTEM Gene among Escherichia Coli Strains Isolated from Different Clinical Samples in Erbil City. 2019;7(1).
2. Mahon CR, Lehman DC, Manuselis G. Textbook of diagnostic microbiology-e-book: Elsevier Health Sciences; 2018.
3. Kalantar D, Mansouri S. Emergence of multiple β-lactamases produced by Escherichia coli clinical isolates from hospitalized patient in Kerman, Iran. 2010.
4. Owaif HAA, Mhawesh AA, Abdulateef SAJAoTM, Health. The role of BipA in the regulation of K1 capsular polysaccharide production of uropathogenic Escherichia coli. 2019;22:201-209.
5. King JE, Owaif HAA, Jia J, Roberts ISJI, immunity. Phenotypic heterogeneity in expression of the K1 polysaccharide capsule of uropathogenic Escherichia coli and downregulation of the capsule genes during growth in urine. 2015;83(7):2605-2613.
6. Drawz SM, Bonomo RAJCmr. Three decades of β-lactamase inhibitors. 2010;23(1):160-201.
7. Palzkill TJAotNYAoS. Metallo-β-lactamase structure and function. 2013;1277:91.
8. Owaif HAHA. Regulation of Transcription of the Escherichia coli Group 2 Capsule Gene Clusters: The University of Manchester (United Kingdom); 2017.
9. Jaber AM, Aal Owaif HJPA. Detection Of Genes Involved In Biofilms Formation By Escherichia Coli Isolated From Patients Suffering Of Urinary Tract Infections. 2020;20(2):5987-5992.
10. Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated Colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. 2016;16(2):161-168.
11. Al-Bdery ASJ, Mohammad GJ, Hussen BJGR. Vancomycin and linezolid resistance among multidrug-resistant Staphylococcus aureus clinical isolates and interaction with neutrophils. 2020;21:100804.
12. Mhawesh AA, Aal Owaif HA, Abdulateef SAJIJoPHR, Development. In vitro Experimental Research for Using the Silver Nanoparticles as Plasmid Curing Agent in Some Types of Multi-Antibiotic Resistant Pathogenic Bacteria. 2019;10(8).
13. Mhawesh AA, Hussein HA, Abdullah MN. GENE EXPRESSION OF MIR-7 IN LOCAL ISOLATES OF STAPHYLOCOCCUS AUREUS.
14. Al-Sa’ady AT, Mohammad GJ, Hussen BMJGR. Genetic relation and virulence factors of Carbapenemase -producing Uropathogenic Escherichia coli from urinary tract infections in Iraq. 2020;21:100911.
15. Ali FA, Hussen BM, Zaki SMJAoTM, Health. Molecular detection of bla ctx-m gene among pseudomonas aeruginosa strains isolated from different clinical samples in erbil city. 2020;23:231-231.
16. Rayamajhi N, Kang SG, Lee DY, et al. Characterization of TEM-, SHV-and AmpC-type β-lactamases from cephalosporin-resistant Enterobacteriaceae isolated from swine. 2008;124(2):183-187.
17. Alkhudhairy MK, Alshadeedi SM, Mahmood SS, Al-Bustan SA, Ghasemian AJMp. Comparison of adhesin genes expression among Klebsiella oxytoca ESBL-non-producers in planktonic and biofilm mode of growth, and imipenem sublethal exposure. 2019;134:103558.
18. Kanaan MHG, Al-Shadeedi SM, Al-Massody AJ, Ghasemian AJCi, microbiology, diseases i. Drug resistance and virulence traits of Acinetobacter baumannii from Turkey and chicken raw meat. 2020;70:101451.
19. Ahmed AM, Nakano H, Shimamoto TJJoAC. The first characterization of extended-spectrum β-lactamase-producing Salmonella in Japan. 2004;54(1):283-284.
20. Balaky STJ, Abdulkhalik H, Hussen BM, Hassan H, Mawlood AHJZJoP, Sciences A. Molecular Identification of Acinetobacter baumanii and Acinetobacter genomic species 13TU Using PCR. 2019;31(1):17-22.
21. Hussen BM, Al-Marzoqi AH, Ghasemian AJJoGC. Assessment of oncogenic role of intestinal microbiota in colorectal cancer patients. 2020:1-6.
22. Hussen BMJZJoP, Sciences A. Evaluating the prevalence of virulence factor gene and biofilm production in Pseudomonas aeruginosa isolated from different clinical samples. 2020;32(4):108-113.
23. Tasli H, Bahar IHJJjoid. Molecular characterization of TEM-and SHV-derived extended-spectrum beta-lactamases in hospital-based Enterobacteriaceae in Turkey. 2005;58(3):162.
24. Villegas MV, Correa A, Perez F, et al. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. 2004;49(3):217-222.
25. Rosenthal VD, Maki DG, Jamulitrat S, et al. International nosocomial infection control consortium (INICC) report, data summary for 2003-2008, issued June 2009. 2010;38(2):95-104. e102.
26. Ling TK, Xiong J, Yu Y, et al. Multicenter antimicrobial susceptibility survey of gram-negative bacteria isolated from patients with community-acquired infections in the People’s Republic of China. 2006;50(1):374-378.
Received: 20 August June 2021 / Accepted: 10 November 2021 / Published: date. 15 February 2022
Citation: Mhawesh A, khudair M, Abbas ON. Major Genetic Determinants of Extended-Spectrum Β-Lactamase (ESBL), Carbapenemase, Fosfomycin and Colistin Resistance in Escherichia Coli from Intensive Care Units. Revis Bionatura 2022;7(1). 11. http://dx.doi.org/10.21931/RB/2022.07.01.11




















