Vol 6 No 4 2021 – 31
REVISION/REVIEW
Study review of camelid and shark antibodies for biomedical and biotechnological applications
Esther Ivanova Matamoros Alcivar; Maily Selena González Avilés,
Available from:http://dx.doi.org/10.21931/RB/2021.06.04.31
ABSTRACT
The antibodies of camelids and sharks are about one–half of the conventional ones while regular antibodies have four protein chains: two light and two heavy, these small antibodies studied have just two heavy chains; they lack a light chain. In recent years, nanobodies have been the focus of attention because they can recognize epitopes that are usually not antigenic (hidden) for conventional antibodies. On the clinical side, researchers are testing nanobodies (Nbs) in the fight against diseases and disease diagnosis. Nanobodies also are attractive because they can prevent protein aggregation and clear the already existing aggregates. Furthermore, new treatments using these Nbs can neutralize the severe acute respiratory syndrome coronavirus (SARS-CoV-2) for preventing COVID-19. In this review, we sum up recent findings of the proposed nanobodies for their potential application.
Keywords: Nanobodies, Camelids, Sharks, Therapeutics, Diseases
INTRODUCTION
Immunoglobulins play a central role in the adaptive immune system by recognizing the specific molecular patterns of external antigens, and through these interactions, they can neutralize and eliminate pathogens from the host.
The three main types of humoral immunoglobulins (Igs) are IgG, IgM, and IgA in the milk of mammals. The concentration of each type of Igs depends on several factors. These factors include the lactation stage and the type of neonatal transfer of antibodies from the mother to the offspring (placental, colostrum or mixed) 1.
While regular antibodies are made of four protein chains (two light and two heavy), the Camelidae family and cartilaginous fishes produce functional antibodies which naturally lack light chains2. These antibodies (Abs) were named single domain antibodies or heavy chain antibodies. Using affinity chromatography techniques performed by Hamers-Casterman3 in 1993, they realized that these Abs have about one–half of the size of the conventional ones.
The heavy chain (VHH) variable region has excellent solubility, stability, tiny size, and a great capacity to recognize unique epitopes, thus having a high specificity and antigen affinity4. It is thought that this super-lightened version of antibodies has evolved independently several times, suggesting that they are highly effective in this animals5.
Immunizing a camelid (camels and llamas) with soluble, adequately folded proteins raises an affinity-matured immune response in the unique camelid heavy-chain only antibodies (HCAbs). The peripheral blood lymphocytes of the immunized animal are used to clone the antigen-binding antibody fragment from the HCAbs in a phage display vector6.
To get these antibodies tailored to a specific molecule, the molecule must first be injected into the animals. Animals then make small antibodies over weeks or months. Researchers use their blood cells for the small antibodies to obtain genes, then use bacteria to produce the antibodies in the laboratory7.
Unlike conventional antibodies, VHH can be cloned, expressed and purified in a prokaryotic system such as E. coli8. The most popular approach to select recombinant antigen-recognizing antibody fragments is the modified panning phage display libraries. This method is based on the selection of DNA sequences that are cloned in a phagemid vector (located within phage particles) and is coding for recombinant proteins, recombinant antibodies, which are able (being expressed as a part of a filamentous phage surface protein on the surface of phage particles) to bind the defined ligand (antigen)7 specifically.
It might soon be easier to obtain nanobodies since a team has made a nanobody library in yeast cells, allowing researchers to skip the animal–inoculation step. iCAN is built as the first comprehensive nanobody database to provide an integrative analysis tool for academic and industrial researchers to expand and accelerate nanobody research2.
Nanobodies and their derivative forms can serve as nano tracers in intracellular bioimaging1 and help visualize the structure of complex proteins due to their nanoscopic size and high affinity against intracellular signaling molecules. They can bind tightly and deeply; this can help stabilize ornate molecules whose flexibility thwarts the imaging process.
On the clinical side, the application of nanobodies in targeting therapeutics shows good prospects in the therapy of acute thrombotic thrombocytopenic purpura9, infectious disease10, rheumatoid arthritis11, central nervous system disease12, breast13 and ovarian cancers14, and so on2. Next on the horizon for the nanobodies is getting them to stick to tumors for imaging purposes15 or using them to allow drugs to cross the blood-brain barrier to tackle diseases of the brain16.
It is essential to know the characteristics of each nanobody to find its proper biomedical or biotechnological applications. This paper review shows the specific characteristics of nanobodies that makes them more potent than the conventional antibodies and will also collect all the advances made in the field of the different species of sharks and camelids’ antibodies, such as the new information collected from ongoing studies of these animals’ immunoglobulins and their potential applications in immunotherapies and fighting diseases.
Methodology
In this study, we critically examined 83 papers published in various journals. The paper selection strategy and evaluation criteria considered in this paper are in Figure 1.
The papers were obtained from Scopus and Google Scholar using keywords’ camelids’, ‘sharks’, ‘nanobodies’, ‘biomedical application’, ‘therapeutic application’, ‘antibodies’, etc.
In this study, we downloaded papers published available online. We found 123 papers and selected 83 papers for review, and filtered out 40 papers that were not useful or not relevant.
In this literature review, 83 papers and 5 books published were critically examined. The information was published in different journals, and some were entrepreneurial web pages (3) since some nanobodies are already in the market.
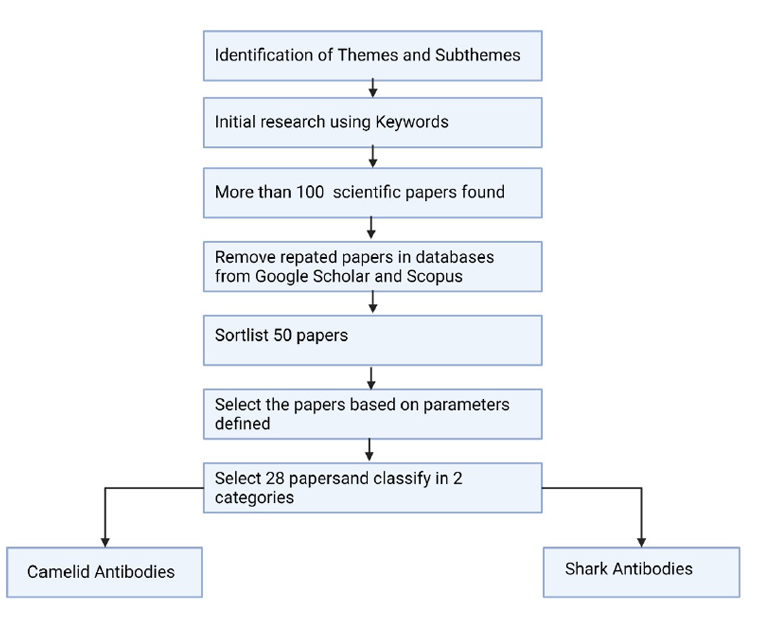
Figure 1. Review methodology for paper selection.
The parameters used for the paper selection were if the study topics such as the nanobody’s therapeutic use, its functional characteristics, biomedical application, biotechnological methods.
The papers selected were separated into two categories: camelid antibodies and shark antibodies.
RESULTS
Camelids antibodies
The camelids of South America are a family of artiodactyl mammals belonging to the order Artiodactyla, suborder Ruminantia, family Camelidae17. The single-domain antibodies are antibodies of the IgG type present in the serum of species of the Camelidae family that lack both the light chain and the CH1 region of the heavy chain18,19.
During pregnancy, some camelids, such as alpacas, cannot transport immunoglobulins from the mother to the fetus, so the offspring are born agammaglobulinemia and require maternal antibodies that will be transferred thanks to colostrum20–22. After the ingestion of colostrum, there is an increase in the IgG concentration in the blood serum of the offspring, which generates resistance to infectious agents23,24.
These antibodies were named single-domain antibodies or heavy chain antibodies because they did not have a light chain, using affinity chromatography techniques performed by Hamers-Casterman3, where the affinity of the antibodies present in the serum of camels for protein A was studied25,26.
Camelids have a single antigen recognition domain so that their unique domain contains a complete antigen-binding site and is the smallest functional antigen-binding fragment (around 15 kDa)27. As is known, antibodies are generally tetramers, composed of 2 dimers called heavy chain (H) of 55 kDa each, and two dimers called light chain (L) of 25 kDa, generally linked by disulfide bridges, each forming a protein in the shape of Y28,29, shown in figure 2.
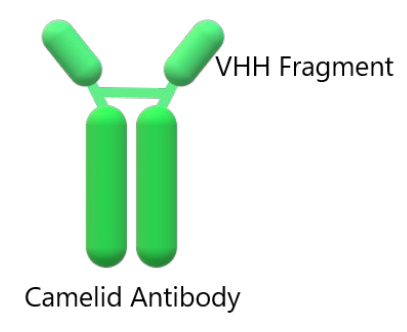
Figure 2. Structure of an IgG camelid immunoglobulin. At the top of this structure is the single-domain antibodies (VHH).
Immunoglobulins of camelids
Currently, these immunoglobulins and especially their simple variable domains (VHHs for short) have various applications in biotechnology29. It has been observed that antigenic sites tend to be more hydrophilic3, flexible, and accessible than the rest of the protein, often protruding from the antigen surface. An early study suggested that conventional Abs have lower surface shape complementarity than other protein-protein complexes30.
Single-domain antibodies or heavy chain antibodies (HCAbs) are G-type immunoglobulins that lack a light chain. In camelids, there are 5 isotypes of antibodies: IgG, IgM, IgA, IgE, and IgD, which differ as much in sequence as in the function of the heavy chain31.
Faced with the invasion of an infectious agent, camelids and other animals activate a humoral response through immunoglobulin G (IgG), preventing the invasion of this infectious agents32. The immunoglobulins, in figure 3, has both the heavy and light chains have a constant region (CH and CL, respectively) and two variable regions (VH and VL, respectively).
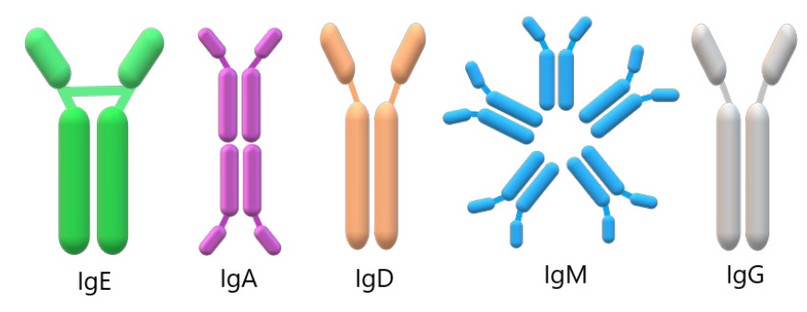
Figure 3. Structure of the types of antibodies found in camelids. According to their H chains, camelids antibodies are classified into five isotypes (IgE, IgA, IgD, IgM, and IgG from left to right), which provide each isotype with distinct characteristics and roles.
Variable region (VH, VL)
The variable region is responsible for recognizing foreign agents or antigens that are molecules that activate the immune system; the amino acid sequence of this region has a significant variability to recognize different antigens33,34.
The variable region of single-domain antibodies, also called VHH, is the minor antibody that can interact with the antigen. It has the advantage of behaving as a monomer independent of the rest of the HCAb; it is very stable and has a low molecular weight of approximately 12-15 kDa. The VHH domain is a fragment of only 15 to 18 KDa35.
Unlike conventional antibodies, VHHs fold independently of the rest of the protein and maintain the ability to have high specificity and affinity for their antigen; they can also be cloned, expressed and purified in a prokaryotic system such as Escherichia coli8.
VHHs are known as nanobodies (Nbs); these Nbs possess properties that allow them to be an ideal tool in research and various therapeutic applications.
Due to these characteristics, VHH has been the object of several studies, mainly for biomedical purposes.
There are many direct applications of VHH in scientific research, for example, as membrane protein stabilizing proteins to facilitate their crystallization34. There is also a significant advance as a therapeutic agent, and even several VHH are in advanced clinical stages for use in humans. However, there are no studies that consider VHH in personalized medicine, such as tissue engineering36.
Constant region (CL, CH)
While the constant domains of the antibodies are not involved in the recognition of antigen. Type G antibodies have three constant domains called CH1, CH2, and CH3 of the heavy chain, and one of the light chains called CL. The IgG is constituted only by heavy chains29.
Potential Biomedical Application of camelids immunoglobulins
The VHH, due to its small size and high stability, has the property of being functional in the interior of living cells and even detecting conformational changes in proteins37,38. In addition to allowing the identification of intracellular elements, VHH can mark proteins for their degradation through the ubiquitination pathway, as the main characteristic of VHH to bind to antigens39,40. This capacity is used in crystallography, where the crystallization chaperones also participate41,42.
The VHH does not require post-translational modifications, so the production of these recombinant VHH is an alternative to monoclonal antibodies for therapeutic use, and they are also very economical and of minimal size with tiny stable molecules43. The nanobodies of camelids have become very popular in cancer treatment since they have a more significant advantage than conventional antibodies44,45. While conventional antibodies are difficult to penetrate and expensive large-scale production, camelid antibodies are more minor, very soluble, stable, easier, and cheaper to produce46.
Studies conducted in mice already show that nanobodies counteract specific tumors in these rodents. Also, these nanobodies have been used in other treatments against life-threatening diseases. More than 85% of human cancers are solid tumours47. In most solid tumors, the necessary first step is to access sufficient amounts of the therapeutic antibody to various tumor regions to obtain a maximal therapeutic effect48. The transfer of macromolecules in tumors is mainly by diffusion, and the speed of diffusion through tumors is inversely proportional to molecular size. In agreement with this, small antibody fragments penetrate the solid tumors more efficiently than the whole antibody molecule49.
Furthermore, to be effective for cancer treatments, nanobodies are also attractive therapeutic options for disorders such as Alzheimer’s and Parkinson’s diseases since they can prevent protein aggregation and, even more interesting, clear the already existing aggregates50. To increase target-to-background signal ratios for high detection sensitivities in imaging and reduce non-specific toxic effects of antibody conjugates, the unbound antibodies should be cleared rapidly from the non-target organs51. When compared to the whole antibody molecule, the small antibody fragments have a faster clearance rate30. The main differences between conventional nanobodies and camelid nanobodies are summarized in table 1.
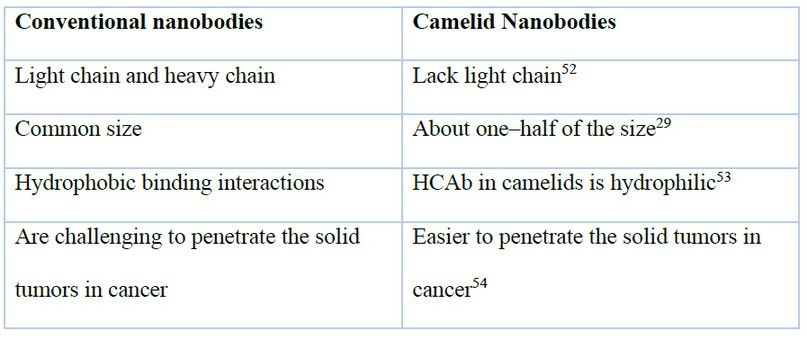
Table 1. Table of main differences between conventional nanobodies and camelid nanobodies
Furthermore, a recent study shows that single-domain antibodies (VHHs) were isolated from a llama immunized with prefusion-stabilized coronavirus spikes. These VHHs neutralize MERS-CoV and SARS-CoV-1 S pseudotyped viruses. The findings provide a molecular basis for VHHs’ ability to neutralize pathogenic beta coronaviruses, implying that these molecules could be effective therapeutics during coronavirus outbreaks10. A summary of this application is in figure 4.
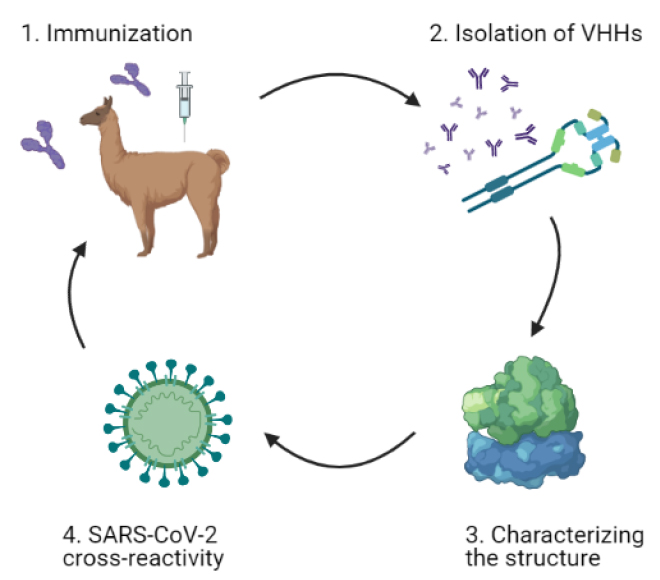
Figure 4. General graphic of single-domain antibodies (VHHs) application in SARS-CoV-2, based on Daniel Wrapp et al., 2020. The schematic representation shows the identified neutralizing cross-reactive single-domain camelid antibodies in llamas immunized with prefusion-stabilized betacoronavirus spike proteins, which may serve as valuable reagents for researchers studying the viruses that cause MERS, SARS, and COVID-1910.
Another study reported by Xiang et al.55 identified a vast repertoire of extremely powerful neutralizing nanobodies (Nbs) to the SARS-CoV-2 spike (S) protein receptor-binding domain (RBD) using camelid immunization and proteomics. They found Nbs that blocked viral infection with picomolar to femtomolar affinity and determined the structure of one of the most potent complexes with RBD. They created multivalent Nb constructs with ultrahigh neutralizing potency (IC50s as low as 0.058 ng/mL) and mutational escape resistance.
Furthermore, a recent study engineered anti-RBD nanobodies from llamas and «nanomice» to produce VHHs cloned from alpacas, dromedaries and camels. They identified two sets of highly neutralizing Nbs using cryo-electron microscopy These findings show that multivalent nanobodies can overcome SARS-CoV-2 variant alterations through two mechanisms: increased avidity for the ACE2 binding domain and identification of conserved epitopes that are difficult to reach with human antibodies. While additional SARS-CoV-2 mutations will continue to arise, nanobodies are promising strategies for preventing COVID-19 mortality if vaccinations are damaged56. This potential alternative of using camelid nanobodies promises to be a revolutionary therapy in searching for new drugs against Coronavirus disease (COVID-19)10,36,57,58.
Cartilaginous fishes antibodies
Sharks are a group of Elasmobranch fish from the class Chondrichthyes or Cartilaginous fish. The evidence of their existence dates back to 450-420 million years ago, placing them in the Ordovician Period59; this makes them the «oldest vertebrate taxon to possess an adaptive immune system» 5 that includes immune factors like immunoglobulins, T cells, Major Histocompatibility Complex (MHC), sites of intense cellular activity, recombinase– activating gene (RAG) activity and also have the earliest version of Ig switching where Somatic Hypermutation (SHM) and Class Switch Recombination (CSR) are contiguous activities60. However, since sharks are cartilaginous species, they do not have bone marrow. Instead, it has been proven that they have an epigonal organ that is the site where the lymphopoiesis occurs, and the B cells are secreted, showing the analogous function of a mammalian bone marrow5.
Cartilaginous fishes immunoglobulins
Sharks have an adaptive immune system based on lymphocyte antigen receptors generated by V(D)J recombinants. Shark B cells express two classical immunoglobulins (Ig), IgM and IgW, encoded by an early, alternative gene organization consisting of numerous autonomous miniloci, where the individual gene cluster carries a few rearranging genes segments and one constant region, µ or ω61. Adult cartilaginous fish express the three immunoglobulins (Ig) isotypes, IgM, IgNAR and IgW. Newborn nurse sharks produce multimeric and monomeric IgM, a germline-joined, IgM-related VH, and meager amounts of monomeric IgM and IgNAR proteins62.
IgW
IgW transcripts, shown in figure 5, are found in secretory full-length, long-form; secretory truncated, short form that is probably derived by alternative splicing63; and trans-membrane long and short forms64.

Figure 5. Schematic representation of the IgW architecture in long and short forms (from left to right)62.
Additionally, a molecular characterization of Ig Heavy chains identified both IgW forms in the African lungfish, Protopterus aethiopicus, a lobe-finned bony fish, which phylogenetic studies suggest are closely related to land vertebrates (tetrapod)63.
The trans-membrane IgW long heavy chain alternative spliced to produce a shorter heavy chain with five domains: one variable and for constant domains, or an even shorter three domains heavy chain: one variable and two constant domains, have been found in the nurse and horn sharks64. And the secreted IgW long heavy chain is composed of seven domains: one variable and six constant domains, and the IgW short heavy chain is composed of three domains: one variable and two constant domains. It is also called IgX or IgNARC64,65. Both IgW secretory forms have also been found in nurse and horn sharks64.
There are multiple-spliced trans-membrane cDNA forms of IgW and IgNAR, possibly selected over evolutionary time to limit proteolysis of the putative cell surface proteins62. There is good evidence of a class switch recombination between IgM and IgW and that IgW(ω) in sharks is orthologous to IgD of vertebrates61,66. IgW short is most highly expressed in the spleen. Also, in unimmunized adults, IgW short transmembrane and secreted forms are produced in the pancreas, suggesting a role in gut mucosal immunity67.
IgM
In B cell receptor systems, the sharks’ IgM exists in both trans-membrane (for activation of lymphocytes) and secretory forms (to bind antigen and induce effector functions of the humoral system)5. This IgM humoral system of sharks was initially thought to be very different from mammals, mainly because the kinetics of the climb in titter was slower to rise than in endotherms67. Serum IgM of immune responsiveness is found in a monomeric 7S and a pentameric (19S) form, both shown in figure 6, present in roughly equal amounts and constituting half of the total serum protein in adult animals 5,65,67. A subclass of the IgM, IgM1gj, was reported in nurse sharks in high amounts as a monomer in the sera of neonates with only three C domains. IgM is expressed in the epigonal organ, but its levels decline in the spleen with age67.
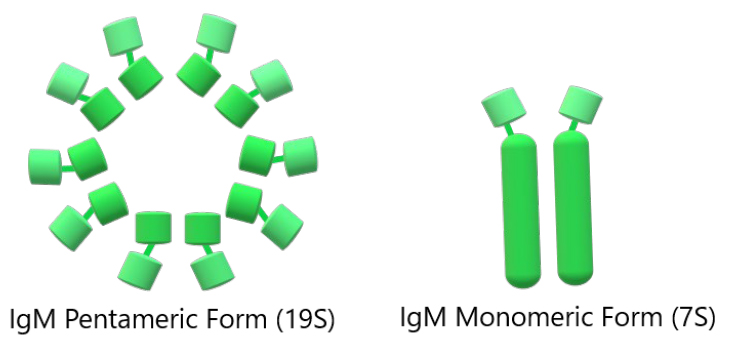
Figure 6. Schematic representation of the overall IgM architecture. The first represents the pentameric form of IgM, and the second represents the monomeric form of IgM (from left to right)60.
Cartilaginous fish express high monomeric and multimeric serum levels of the same Ig isotype65 in approximately equal amounts despite independent production from the same set of IgH loci 67 26. Immunoglobulins of the IgM class generally have disulfide-linked pentamers of total molecular weight near 900000. Each monomer unit is similar to an IgG molecule, containing two light (L) and two heavy chains linked by disulfide bonds and noncovalent interactions68.
IgM has been found in the eyes, gills, intestine, liver, pancreas, peripheral blood leukocytes, spleen and testis67.
IgNAR
Surface Ig New Antigen Receptors (IgNARs) are formed by two identical heavy chains composed of five or three constant domains, shown in figure 6, a variable domain (V-NAR) and one amino-terminal V domain 65,66. The first and third constant domains homodimerize and lend to the antibody stability, and the resolution of the stable region structure showed stabilizing motifs 66,69.
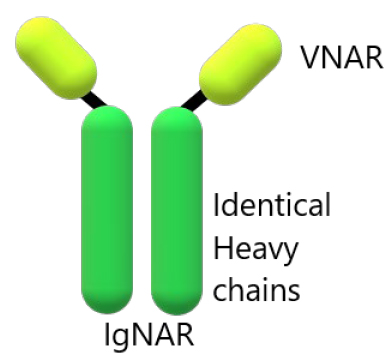
Figure 7. Schematic representation of the overall IgNAR architecture. Shark antibody-containing identical heavy chains 69,70.
The IgNAR is quite an unconventional antibody being a homodimer of H chains that do not associate with L chains71, and since it does not fit the profile of an actual antibody although it is structurally similar5. The variability within the CDR1 and CDR3 regions69 certainly suggested so, as did the general organization into a variable-constant domain format and the occurrence of transmembrane and secretory forms of the protein72. Nevertheless, it follows an analogous target–specific kinetics of expression in response to an immunogenic challenge 5,73.
The IgNARV is the smallest antigen-binding domain known in the animal kingdom and is also particularly water– soluble66,74. VNARs are classified into types based upon the number and location of additional noncanonical cysteine (cys) residues in their variable domain. These cys form interdomain disulfide bonds that dictate the structure of the VNAR domain 71,72. Type I VNARs have germline-encoded, noncanonical cys residues in framework regions (FR)2 and FR4, that pair with D region encoded cys residues in CDR3; this folds the long CDR3 loop over the surface that would associate with VL in conventional Igs, pinning it there.
In contrast, type II VNARs either have a single germline-encoded, noncanonical cys residue in CDR1 that pairs with a lone cys present in CDR3, forming a stabilizing disulfide bond between the protruding antigen-binding loops, or no extra cys (sometimes called type IIb or type IV) allowing CDR3 to move away from CDR1. Type I CDR3’s are longer on average than those of type II, undoubtedly a result of selection due to their unique disulfide bonding patterns.
A third VNAR type, type III, so far found only in nurse sharks, has a structure very similar to type II VNARS; however, CDR3 diversity in this type is drastically reduced due to germline fusion of two (D1 and D2) of the three D segments present in this cluster71. There is a limited sequence diversity of type III CDRs structural modeling; however, it has been revealed invariant stabilizing aromatic residues in CDR1 (tryptophan 31) and CDR3 (phenylalanine 96), which could allow a more excellent range of structural diversity than would be predicted for this VNAR type. Expressed early in development and retaining expression only in the epigonal of adult sharks, type III is hypothesized to mediate the first line of immune defense in neonates; however, the antigens it targets still require investigation73.
Much remains to be understood about the role of IgNAR in the immune system of cartilaginous fishes. So far, isolation of antigen-specific VNARs from libraries has primarily been conducted via bacteriophage (phage) display21. In this case, the library of VNAR sequences is cloned in-frame with a phage coat protein (usually the gene III protein) encoded in a phagemid vector. However, it is abundantly clear that VNARs present a singular molecular framework that can be exploited to develop reagents for scientific research, disease diagnosis, and/or potentially therapeutic intervention 65,72.
Potential Biomedical Applications of Cartilaginous fishes immunoglobulins
The IgW is an Ig isotype that exists in two forms, one of full length and one lacking the Fc region61. This isotype can bind antigen without evoking Fc-mediated responses, thus limiting inflammation65. The possibility of an anti-IgW antibody is being investigated that will provide an essential research tool to studying cartilaginous fishes’ adaptive immune system75. Sharks can produce an IgM-based response following immunization, evidence for memory and affinity maturation65.
IgM might have a role in blood osmoregulation76 analogous to the physiology of albumin in other vertebrates and have been shown to be active in lytic, opsonic and antibody-induced cytotoxicity-like reactions77. This killing is observed through phagocytosis, which is mediated by both 7S and 19S IgM antibodies. Throughout evolution, IgM reacts with particulate antigen and through binding Fc– receptors on the surface of leukocytes, enhancing phagocytosis67.
The relative stability of the IgNAR heavy chain homodimer, the extraordinary stability and resistance to irreversible denaturation of VNAR domains74 and the somewhat simpler genetics encoding the single variable domain have fuelled applied research to adapt this molecule to biotechnology and immunotherapy66. IgNAR has a small structure and an excellent binding ability. Its single variable domain represents an opportunity to bind different epitopes than traditional antibodies and can also work as an autonomous paratope74.
For diseases like protein aggregation disorders such as Alzheimer78 and Parkinson79, nanobodies can prevent protein aggregation and, even more interestingly, clear the already existing aggregates50. Ossianix Inc80 has utilized transferrin receptors to demonstrate that they can transfer VNAR antibodies across the blood-brain barrier81 shown in figure 7.
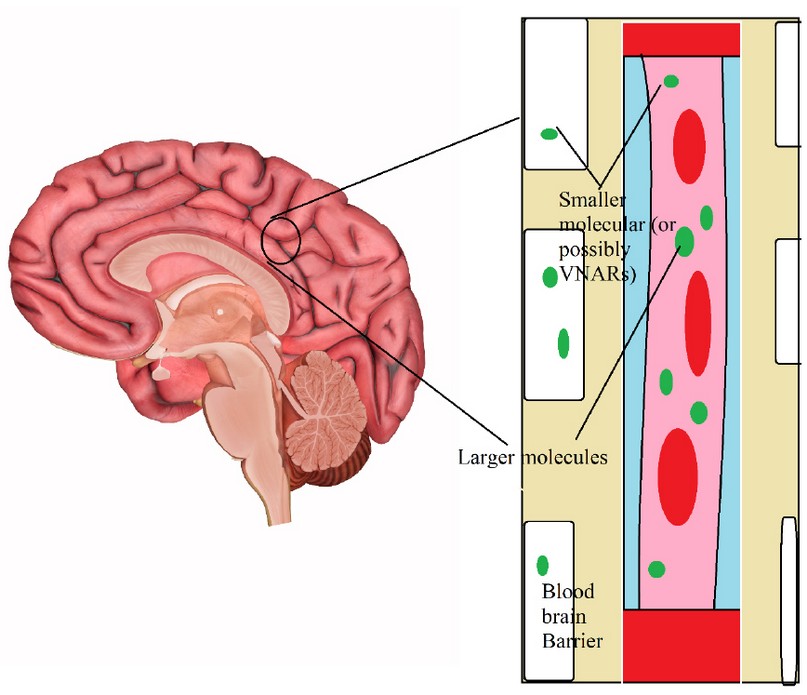
Figure 8. Mechanism of therapeutic use for nanobodies in neurological diseases. Nanobodies can pass the blood-brain barrier74.
In addition, they are also developing VNAR-therapies for neurological, neuromuscular as well as autoimmune diseases 74,80. Antigen-specific VNAR molecules have been isolated against a wide range of disease–related targets, including proteins involved in cancer82 and arthritis83, cytokines84, toxins85, and viral targets. However, with their small size, high stability, and possible advantages in cryptic epitope recognition, VNARs are natural candidates for future niche biotechnological and diagnostic applications 71,72.
There has been a recent study73 to evaluate the VNARs’ potential for therapeutic development, where researchers chose a variety of human tumor biomarkers and virus antigen proteins as selection targets, including glypican-3 (GPC3), HER2 and PD1, the spike proteins of the MERS and SARS viruses, and Pseudomonas exotoxin (PE38). They developed a method to construct phage display libraries based on PCR extension assembly followed by self-ligation. The selected binders, Type I and II VNARs, were produced successfully in E. coli as soluble proteins for antigen-binding validation. This study validates the diversity of the nurse shark VNAR library and the utility of the shark VNAR library as a platform for therapeutic antibody discovery.
A nave wobbegong VNAR library was used to identify two natural clones of micromolar (M) affinity for the protease gingipain K (Kgp), a virulence factor of Porphyromonas gingivalis, which causes periodontal disease in humans71. Researchers used panning a semisynthetic wobbegong shark library to isolate a VNAR capable of targeting Hepatitis B virus (HBV) pre-core protein, which is secreted as Hepatitis B e antigen (HBeAg)71. It also may work as half-life extension tools in disease treatment74. Engineering stability into a shark VNAR combined strategies of CDR grafting and consensus sequences mutagenesis have been developed in a VNAR specific for the nucleoprotein of the Ebola virus86.
Recently, a group of researchers tested the VNAR domains for the treatment of COVID-19. They used the neutralizing properties of the VNAR against the Spike protein from the SARS-CoV-2 Wuhan variant. The antibodies showed great affinity and high blocking of the ACE2 receptors’ interaction with the virus variant. This study expands the alternative treatments against the new virus87.
To produce VNARs for the above applications, individual clones that bind the target of interest with high specificity and good affinity must first be isolated from an extensive repertoire of diverse sequences. Two strategies have been utilized to generate natural VNAR libraries: 1) amplifying the VNAR repertoire from one or more naive animals or 2) from animals immunized with the target antigen71. The application of each immunoglobulin explained in this review is summed up in table 2.
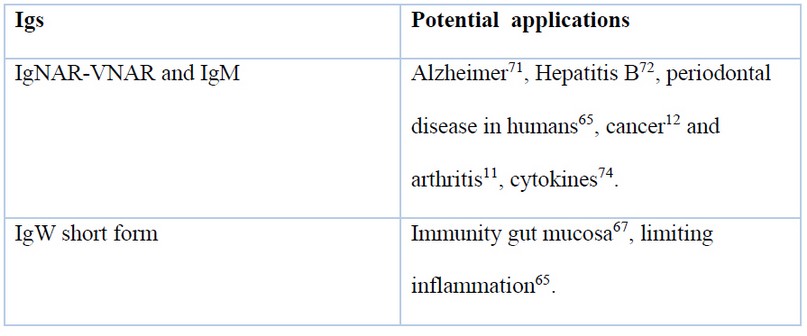
Table 2. Table of Sharks immunoglobulins and their application
CONCLUSIONS
As presented previously, the nanobodies of the camelids not only have characteristics that have made them very popular for cancer treatments, but they are also desirable therapeutic options for disorders such as Alzheimer’s disease and Parkinson’s disease. These studies in sharks and camelids can also be performed in Ecuador since these animals live within the fauna of this biodiverse country and can boost biomedical and biotechnological research to get better and effective therapies and treatments for better and effective therapies and treatments in hospitals and clinics. Nanobodies have reached many publications that are increasing in recent years; one of the fields very interested in these investigations of nanobodies are the large pharmaceutical industries, which reflect high expectations for nanoantibody technology.
Despite the exhaustive study of the sharks’ immunoglobulins, there is still a lack of information about the possible applications of the recently deeply studied immunoglobulins IgW, IgM and IgNAR. Although their characteristics seem very promising on the clinical side, especially in the therapeutic and antiviral treatments, few experiments have not gotten further than mice experimentation. For example, while VNARs have been raised against a diverse array of targets, most studies have confirmed antigen specificity, affinity, and stability; very few studies have proceeded as far as testing the VNARs in relevant disease models. Further work is needed to establish the structural and functional repertoire of VNAR domains and their potential immunogenicity.
REFERENCES
1 Leduc C, Si S, Gautier JJ, Gao Z, Shibu ES, Gautreau A et al. Single-molecule imaging in live cell using gold nanoparticles. Methods Cell Biol 2015; 125: 13–27.
2 Zuo J, Li J, Zhang R, Xu L, Chen H, Jia X et al. Institute collection and analysis of Nanobodies (iCAN): a comprehensive database and analysis platform for nanobodies. BMC Genomics 2017; 18: 797.
3 Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hammers C, Songa EB et al. Naturally occurring antibodies devoid of light chains. Nature 1993; 363: 446–448.
4 Rabia LA, Desai AA, Jhajj HS, Tessier PM. Understanding and overcoming trade-offs between antibody affinity, specificity, stability and solubility. Biochem Eng J 2018; 137: 365–374.
5 Barelle C, Porter A. VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binder of Proteins. Antibodies 2015; : 240–258.
6 Vincke C, Gutiérrez C, Wernery U, Devoogdt N, Hassanzadeh-Ghassabeh G, Muyldermans S. Generation of Single Domain Antibody Fragments Derived from Camelids and Generation of Manifold Constructs. In: Chames P (ed). Antibody Engineering: Methods and Protocols, Second Edition. Humana Press: Totowa, NJ, 2012, pp 145–176.
7 Brissette R, Goldstein NI. The Use of Phage Display Peptide Libraries for Basic and Translational Research. In: Fisher PB (ed). Cancer Genomics and Proteomics: Methods and Protocols. Humana Press: Totowa, NJ, 2007, pp 203–213.
8 Tanha J, Dubuc G, Hirama T, Narang SA, MacKenzie CR. Selection by phage display of llama conventional VH fragments with heavy chain antibody VHH properties. J Immunol Methods 2002; 263: 97–109.
9 Holz J-B. The TITAN trial – Assessing the efficacy and safety of an anti-von Willebrand factor nanobody in patients with acquired thrombotic thrombocytopenic purpura. Transfus Apher Sci 2012; 46: 343–346.
10 Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020; 181: 1004-1015.e15.
11 Put S, Schoonooghe S, Devoogdt N, Schurgers E, Avau A, Mitera T et al. SPECT Imaging of Joint Inflammation with Nanobodies Targeting the Macrophage Mannose Receptor in a Mouse Model for Rheumatoid Arthritis. J Nucl Med 2013; 54: 807–814.
12 Demeules M, Scarpitta A, Abad C, Gondé H, Hardet R, Pinto-Espinoza C et al. Evaluation of P2X7 Receptor Function in Tumor Contexts Using rAAV Vector and Nanobodies (AAVnano). Front Oncol 2020; 10: 1699.
13 Altunay B, Morgenroth A, Beheshti M, Vogg A, Wong NCL, Ting HH et al. HER2-directed antibodies, affibodies and nanobodies as drug-delivery vehicles in breast cancer with a specific focus on radioimmunotherapy and radioimmunoimaging. Eur J Nucl Med Mol Imaging 2021; 48: 1371–1389.
14 Vaneycken I, Devoogdt N, Van Gassen N, Vincke C, Xavier C, Wernery U et al. Preclinical screening of anti-HER2 nanobodies for molecular imaging of breast cancer. FASEB J 2011; 25: 2433–2446.
15 Keyaerts M, Xavier C, Everaert H, Vaneycken I, Fontaine C, Decoster L et al. Phase II trial of HER2-PET/CT using 68Ga-anti-HER2 VHH1 for characterization of HER2 presence in brain metastases of breast cancer patients. Ann Oncol 2019; 30: iii25–iii26.
16 Bélanger K, Iqbal U, Tanha J, MacKenzie R, Moreno M, Stanimirovic D. Single-Domain Antibodies as Therapeutic and Imaging Agents for the Treatment of CNS Diseases. Antibodies 2019; 8. doi:10.3390/antib8020027.
17 Fowler ME. Camelids Are Not Ruminants. Zoo Wild Anim. Med. 2008; : 375–385.
18 Liu JL, Zabetakis D, Brown JC, Anderson GP, Goldman ER. Thermal stability and refolding capability of shark derived single domain antibodies. Mol Immunol 2014; 59: 194–199.
19 Könning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol 2017; 45: 10–16.
20 Daley-Bauer LP, Purdy SR, Smith MC, Gagliardo LF, Davis WC, Appleton JA. Contributions of conventional and heavy-chain IgG to immunity in fetal, neonatal, and adult alpacas. Clin Vaccine Immunol 2010; 17: 2007–2015.
21 Majewska M, Panasiewicz G, Szafranska B. Expression of pregnancy-associated glycoprotein family in the epitheliochorial placenta of two Camelidae species (C. dromedarius and C. bactrianus). Acta Histochem 2013; 115: 669–676.
22 Skidmore JA. The use of some assisted reproductive technologies in old world camelids. Anim Reprod Sci 2019; 207: 138–145.
23 Garmendia AE, Palmer GH, DeMartini JC, McGuire TC. Failure of passive immunoglobulin transfer: a major determinant of mortality in newborn alpacas (Lama pacos). Am J Vet Res 1987; 48: 1472–1476.
24 Paul-Murphy J. Obstetrics, Neonatal Care, and Congenital Conditions. Vet Clin North Am Food Anim Pract 1989; 5: 183–202.
25 Arbabi-Ghahroudi M. Camelid single-domain antibodies: Historical perspective and future outlook. Front Immunol 2017; 8. doi:10.3389/fimmu.2017.01589.
26 Gonzalez-Sapienza G, Rossotti MA, Tabares-da Rosa S. Single-Domain Antibodies As Versatile Affinity Reagents for Analytical and Diagnostic Applications. Front Immunol 2017; 8: 977.
27 Stanimirovic D, Kemmerich K, Haqqani AS, Farrington GK. Engineering and pharmacology of blood-brain barrier-permeable bispecific antibodies. Adv Pharmacol 2014; 71: 301–335.
28 Zavrtanik U, Lukan J, Loris R, Lah J, Hadži S. Structural Basis of Epitope Recognition by Heavy-Chain Camelid Antibodies. J Mol Biol 2018; 430: 4369–4386.
29 Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007; 77: 13–22.
30 Shire SJ, Gombotz W, Bechtold-Peters K, Andya J. Current trends in monoclonal antibody development and manufacturing. Springer Science \& Business Media, 2009.
31 Wernery U. Camelid Immunoglobulins and Their Importance for the Newborn – A Review. J Vet Med Ser B 2001; 48: 561–568.
32 De Simone EA, Saccodossi N, Ferrari A, Leoni J. Development of ELISAs for the measurement of IgM and IgG subclasses in sera from llamas (Lama glama) and assessment of the humoral immune response against different antigens. Vet Immunol Immunopathol 2008; 126: 64–73.
33 Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett 1997; 414: 521–526.
34 Desmyter A, Transue TR, Ghahroudi MA, Dao Thi M-H, Poortmans F, Hamers R et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol 1996; 3: 803–811.
35 Bannas P, Hambach J, Koch-Nolte F. Nanobodies and Nanobody-Based Human Heavy Chain Antibodies As Antitumor Therapeutics. Front Immunol 2017; 8: 1603.
36 Bessalah S, Jebahi S, Mejri N, Salhi I, Khorchani T, Hammadi M. Perspective on therapeutic and diagnostic potential of camel nanobodies for coronavirus disease-19 (COVID-19). 3 Biotech 2021; 11: 89.
37 Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LGJ et al. Single-domain antibody fragments with high conformational stability. Protein Sci 2002; 11: 500–515.
38 Rajan M, Mortusewicz O, Rothbauer U, Hastert FD, Schmidthals K, Rapp A et al. Generation of an alpaca-derived nanobody recognizing γ-H2AX. FEBS Open Bio 2015; 5: 779–788.
39 Staus DP, Wingler LM, Strachan RT, Rasmussen SGF, Pardon E, Ahn S et al. Regulation of β2-Adrenergic Receptor Function by Conformationally Selective Single-Domain Intrabodies. Mol Pharmacol 2014; 85: 472–481.
40 Desmyter A, Spinelli S, Roussel A, Cambillau C. Camelid nanobodies: killing two birds with one stone. Curr Opin Struct Biol 2015; 32: 1–8.
41 Böldicke T. Single domain antibodies for the knockdown of cytosolic and nuclear proteins. Protein Sci 2017; : 40.
42 Traenkle B, Emele F, Anton R, Poetz O, Haeussler RS, Maier J et al. Monitoring Interactions and Dynamics of Endogenous Beta-catenin With Intracellular Nanobodies in Living Cells *[S]. Mol Cell Proteomics 2015; 14: 707–723.
43 Soetens E, Ballegeer M, Saelens X. An Inside Job: Applications of Intracellular Single Domain Antibodies. Biomolecules 2020; 10. doi:10.3390/biom10121663.
44 Unciti-Broceta JD, Del Castillo T, Soriano M, Magez S, Garcia-Salcedo JA. Novel therapy based on camelid nanobodies. Ther Deliv 2013; 4: 1321–1336.
45 Yang EY, Shah K. Nanobodies: Next Generation of Cancer Diagnostics and Therapeutics. Front Oncol 2020; 10: 1182.
46 Siontorou CG. Nanobodies as novel agents for disease diagnosis and therapy. Int J Nanomedicine 2013; 8: 4215–4227.
47 Al-Zu’bi M, Mohan A. Modelling of combination therapy using implantable anticancer drug delivery with thermal ablation in solid tumor. Sci Rep 2020; 10: 19366.
48 Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer 2012; 12: 278–287.
49 Cortez-Retamozo V, Lauwereys M, Hassanzadeh Gh G, Gobert M, Conrath K, Muyldermans S et al. Efficient tumor targeting by single-domain antibody fragments of camels. Int J cancer 2002; 98: 456–462.
50 Ghassabeh GH, Muyldermans S, Saerens D. Nanobodies, Single-Domain Antigen-Binding Fragments of Camelid Heavy-Chain Antibodies. In: Shire SJ, Gombotz W, Bechtold-Peters K, Andya J (eds). Current Trends in Monoclonal Antibody Development and Manufacturing. Springer New York: New York, NY, 2010, pp 29–48.
51 Khaleghi S, Khoshtinat Nikkhoi S, Rahbarizadeh F. Camelid Single-Domain Antibodies for Targeting Cancer Nanotheranostics. In: Saravanan M, Barabadi H (eds). Cancer Nanotheranostics: Volume 1. Springer International Publishing: Cham, 2021, pp 93–123.
52 Vincke C, Muyldermans S. Introduction to Heavy Chain Antibodies and Derived Nanobodies. In: Saerens D, Muyldermans S (eds). Single Domain Antibodies: Methods and Protocols. Humana Press: Totowa, NJ, 2012, pp 15–26.
53 Eggers M, Rühl F, Haag F, Koch-Nolte F. Nanobodies as probes to investigate purinergic signaling. Biochem Pharmacol 2021; 187: 114394.
54 Kolkman JA, Law DA. Nanobodies – from llamas to therapeutic proteins. Drug Discov Today Technol 2010; 7: e139–e146.
55 Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science (80- ) 2020; 370: 1479–1484.
56 Xu J, Xu K, Jung S, Conte A, Lieberman J, Muecksch F et al. Multimeric nanobodies from camelid engineered mice and llamas potently neutralize SARS-CoV-2 variants. bioRxiv 2021. doi:10.1101/2021.03.04.433768.
57 Dubey A, Dahiya S, Rouse BT, Sehrawat S. Perspective: Reducing SARS-CoV2 Infectivity and Its Associated Immunopathology. Front Immunol 2020; 11: 581076.
58 Huo J, Mikolajek H, Le Bas A, Clark JJ, Sharma P, Kipar A et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat Commun 2021; 12: 5469.
59 Martin A. Biology of Sharks and Rays. ReefQuest Cent. Shark Res. 2006.http://www.elasmo-research.org/education/evolution/geologic_time.htm.
60 Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L et al. Origin of immunoglobulin isotype switching. Curr Biol 2012; 22: 872–880.
61 Zhang C, Du Pasquier L, Hsu E. Shark IgW C Region Diversification through RNA Processing and Isotype Switching. J Immunol 2013; 191: 3410–3418.
62 Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented Multiplicity of Ig Transmembrane and Secretory mRNA Forms in the Cartilaginous Fish. J Immunol 2004; 173: 1129–1139.
63 Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol 2004; 5: 1–15.
64 Smith SL, Sim RB, Flajnik MF. Immunobiology of the Shark. CRC Press, 2014.
65 Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol 2005; 35: 936–945.
66 de los Rios M, Criscitiello MF, Smider V V. Structural and genetic diversity in antibody repertoires from diverse species. Curr Opin Struct Biol 2015; 33: 27–41.
67 Mashoof S, Criscitiello MF. Fish Immunoglobulins. Biology (Basel) 2016; 5. doi:10.3390/biology5040045.
68 Lakowicz JR. Topics in Fluorescence Spectroscopy: Principles. Springer Science \& Business Media, 2006.
69 Cheong WS, Leow CY, Abdul Majeed AB, Leow CH. Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody. Int J Biol Macromol 2020; 147: 369–375.
70 Griffiths K, Dolezal O, Parisi K, Angerosa J, Dogovski C, Barraclough M et al. Shark Variable New Antigen Receptor (VNAR) Single Domain Antibody Fragments: Stability and Diagnostic Applications. Antibodies 2013; 2: 66–81.
71 Matz H, Dooley H. Shark IgNAR-derived binding domains as potential diagnostic and therapeutic agents. Dev Comp Immunol 2019; 90: 100–107.
72 Nuttall SD, Krishnan U V, Hattarki M, De Gori R, Irving RA, Hudson PJ. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol Immunol 2001; 38: 313–326.
73 Feng M, Bian H, Wu X, Fu T, Fu Y, Hong J et al. Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naïve nurse sharks. Antib Ther 2018; 2: 1–11.
74 Könning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017; 45: 10–16.
75 Ayham A. Anti-IgW [V42] | Monoclonal Antibodies. Vertebr. Antibodies Ltd. https://ximbio.com/reagent/153310/anti-igw-v42#references (accessed 1 Nov2021).
76 Barelle C, Gill DS, Charlton K. Shark Novel Antigen Receptors—The Next Generation of Biologic Therapeutics? In: Guzmán CA, Feuerstein GZ (eds). Pharmaceutical Biotechnology. Springer New York: New York, NY, 2009, pp 49–62.
77 McKinney EC, Flajnik MF. IgM-mediated opsonization and cytotoxicity in the shark. J Leukoc Biol 1997; 61: 141–146.
78 Meier SR, Sehlin D, Hultqvist G, Syvänen S. Pinpointing Brain TREM2 Levels in Two Mouse Models of Alzheimer’s Disease. Mol Imaging Biol 2021; 23: 665–675.
79 Sehlin D, Stocki P, Gustavsson T, Hultqvist G, Walsh FS, Rutkowski JL et al. Brain delivery of biologics using a cross-species reactive transferrin receptor 1 VNAR shuttle. FASEB J 2020; 34: 13272–13283.
80 Ossianix. Ossianix – a single domain antibody company. 2021.https://www.ossianix.co.uk/ (accessed 1 Nov2021).
81 Vaughan T, Osbourn J, Jallal B, Mannhold R, Folkers G, Buschmann H. Protein Therapeutics, 2 Volume Set. Wiley, 2017https://books.google.com.ec/books?id=3tYtDwAAQBAJ.
82 Walsh FS, Wicher K, Szary J, Stocki P, Demydchuk M, Rutkowski L. Abstract 3631: Delivery of a CD20 transferrin receptor VNAR bispecific antibody to the brain for CNS lymphoma. Cancer Res 2017; 77: 3631.
83 Ubah OC, Steven J, Porter AJ, Barelle CJ. An Anti-hTNF-α Variable New Antigen Receptor Format Demonstrates Superior in vivo Preclinical Efficacy to Humira® in a Transgenic Mouse Autoimmune Polyarthritis Disease Model. Front Immunol 2019; 10: 526.
84 Husain B, Ellerman D. Expanding the Boundaries of Biotherapeutics with Bispecific Antibodies. BioDrugs 2018; 32: 441–464.
85 Liu JL, Anderson GP, Delehanty JB, Baumann R, Hayhurst A, Goldman ER. Selection of cholera toxin specific IgNAR single-domain antibodies from a naïve shark library. Mol Immunol 2007; 44: 1775–1783.
86 Goldman ER, Liu JL, Zabetakis D, Anderson GP. Enhancing stability of camelid and shark single domain antibodies: An overview. Front Immunol 2017; 8: 1–11.
87 Gauhar A, Privezentzev C V, Demydchuk M, Gerlza T, Rieger J, Kungl AJ et al. Single domain shark VNAR antibodies neutralize SARS-CoV-2 infection in vitro. bioRxiv 2021. doi:10.1101/2021.06.08.447530.
88 Tillib S V. «Camel nanoantibody» is an efficient tool for research, diagnostics and therapy. Mol Biol 2011; 45: 66–73.
Received: 24 July 2021
Accepted: 31 October 2021
Esther, Ivanova, Matamoros, Alcivar, orcid.org/0000-0002-5422-630X]1
Maily, Selena, González, Avilés, orcid.org/0000-0001-9399-6940]1*
1School of Biology Science and Engineering, Yachay Tech University of Technology and Research, Ecuador
*Corresponding author: Maily Selena González Avilés maily.gonzalez@yachaytech.edu.ec



















