Vol 6 No 3 2021 – 11
INVESTIGATION / RESEARCH
Tips for a reduction of false positives in manual RT-PCR diagnostics of SARS-CoV-2
Francisco J. Alvarez, Mariela Perez-Cardenas, Marco Gudiño, Markus P. Tellkamp
Available from: http://dx.doi.org/10.21931/RB/2021.06.03.11
ABSTRACT
RT-PCR is the standard gold technique for testing the presence of RNA of the coronavirus causing Severe Acute Respiratory Syndrome (SARS-CoV-2) due to its high specificity and sensitivity. Despite its general use and reliability, no lab in the world is immune to the generation of false positives. These errors cause a loss of confidence in the technique’s power and damage the image of laboratories. More importantly, they can take a toll on tested individuals and have economic, psychological, and health-associated effects. Most false positives are caused during a manual operation inside the laboratory. However, not much has been published about the errors associated with particular laboratory techniques used to detect the virus since the beginning of the actual pandemic. This work precisely reflects on events that occur during manual RT-PCR diagnostics in a COVID-19 laboratory, providing tips for reducing false-positive results.
Keywords: SARS-CoV-2; false positives; RNA extraction; RT-PCR
INTRODUCTION
In COVID-19 diagnostics labs, most errors in reporting consist of false negatives due to the low viral load that escapes detection. Therefore, the sampling time is significant: a sample taken too early after the person has been infected may have too low a viral load to be detected1. Likewise, a sample taken after the patient has cleared the virus almost wholly would result in a very low viral titer. The skills of the doctor or nurse in charge of taking the samples from patients can also contribute to generating false negatives. In addition, the choice of transport medium where the sample is carried to the labs for analysis, whether the sample contains blood, and the temperature during storage and/or transportation can make a difference in the quality of the sample before its processing. False negatives are of great concern for public health management, but the impact of false positives has come to the fore recently due to the distress they can cause in the lives of patients and public health.
The RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) is the standard gold technique for detecting the SARS-CoV-2 virus. It is routinely used for samples from diverse origins, such as nasopharyngeal and throat swabs, sputum, broncho-alveolar lavages2, and anal swabs3. It can detect as few as 5 copies of the virus in a sample4 and is, therefore, the technique of choice to reveal which individuals are contagious at any time of their infection period. They outcompete serological tests, which need a higher viral load to produce a positive result or indirectly measure the virus’s presence in the organism. The RT-PCR has a sensitivity (ability to detect true positives) of 70% and a specificity (ability to report well true negatives) close to 100%5,6. Having such sensitivity, the technique can easily amplify contaminating virus particles that don’t belong to the actual samples to be tested. Hence, RT-PCR can generate false positives with a significant likelihood. Although few studies report on rates of false positives for COVID-19, some estimates of false-positive rates suggest them to be in a range of about 0.3-4%7,8,9. For SARS-CoV and MERS-CoV, the rate oscillated between 0.3-6.9%10 .
False positives can also arise due to errors in reporting the results and uncertainties regarding the cycle-threshold (Ct) value used as a diagnostic criterium11. The former point can be addressed by judicious data entry; however, the latter poses a true challenge when Ct values are close to the cut-off. Here, the likelihood of a false positive or false negative is highest. For instance, if a Ct of 40 is chosen as the diagnostic criterium, does a Ct of 39.5 necessarily mean that 1) the true Ct is indeed below 40 and 2) is the patient still infectious?11,12
A false positive report on healthy people can have dramatic consequences, varied and challenging to quantify. A politician may be deprived of a critical meeting for the citizens he represents. A skilled celebrity may be deprived of a significant sports competition. Worse even, the economic distress caused to those belonging to low-income groups in the society is more significant, for they may have to stop working, may have dependents, and no savings. The stigma of being called positive and the fear of suffering complications can also be detrimental psychologically for many. Besides, healthy but wrongly diagnosed positive, people may be put at risk of real contagion when moved to areas in a hospital with infected patients or may suffer the delay of an essential medical procedure10,13. Unfortunately, due to the severity of the current pandemic, it is not feasible to perform confirmatory PCRs for every patient whose sample yielded a positive result. Instead, current guidelines by international and national public health agencies recommend evaluating every PCR result on a case by case basis in combination with the evaluation of local infection rates, clinical signs and symptoms, lung CT scans, and history of exposure10. The consequences for the labs that report false positives and negatives can also be dramatic. With their image damaged, contracts for private labs may be postponed, and potential customers’ confidence can vanish. Public labs are also questioned when reports of false positives appear. However, false positives are unavoidable and cannot be eliminated in any laboratory. However, it should be possible to reduce them to a minimum to better comply with the targets of national regulatory agencies.
The standard laboratory workflow for COVID-19 testing goes in one direction and has multiple barriers to prevent cross-contamination. Laboratory technicians have routinely tested themselves for covid, for apparent reasons. Still, the laboratory is the primary source of false positives, mainly cross-contamination due to human error10. In this review, we’ll discuss working habits that reduce the occurrence of false-positive reports of coronavirus infection while navigating the manual processing of nasopharyngeal swabs in all phases of the workflow. The tips provided are based on personal experience of false positives while working in a COVID-19 test lab at Yachay Tech University, using typical kits and equipment.
Reduction of false positives in the RNA extraction area
The manual processing of samples during RNA extraction constitutes the highest source of false positives in the laboratory, but there is much room for improvement. The processing of samples in the RNA extraction area starts with setting the laminar flow hood under UV light for several minutes while the airflow stabilizes14. A small centrifuge should be located inside the hood, and it should be left open during UV irradiation to expose the rotor. The space under the hood should not be crowded to help the airflow exert its function and allow the UV light to reach most surfaces. If more than one batch of samples is going to be processed in the day, it is advisable to have rounds of UV irradiation in between batches. Next, parts and equipment to be used to process the samples are cleaned with 70% ethanol. Only filtered tips are used for the extraction and must be changed for every sampling step of the extraction process. Tube racks should be stable to avoid spills. Reagents should be aliquots of the original kit contents to minimize costs if contamination (reagent contamination is a significant source of false positives but easy to identify because most likely, all samples of the same batch would come out positive and with similar Ct values). Lab technicians must wear protective gear with very fit pairs of gloves, without folds on the surface of their fingertips. Once the site is clean, the lab technician starts preparing a master mix that ensures that all the samples get the same initial buffer solution. Fig. 1 shows the process of RNA extraction for a typical commercial kit, with few modifications of the original protocol. Similar steps apply to many other commercial kits. Double arrows A to C at the bottom indicate the times at which different contamination types can occur.
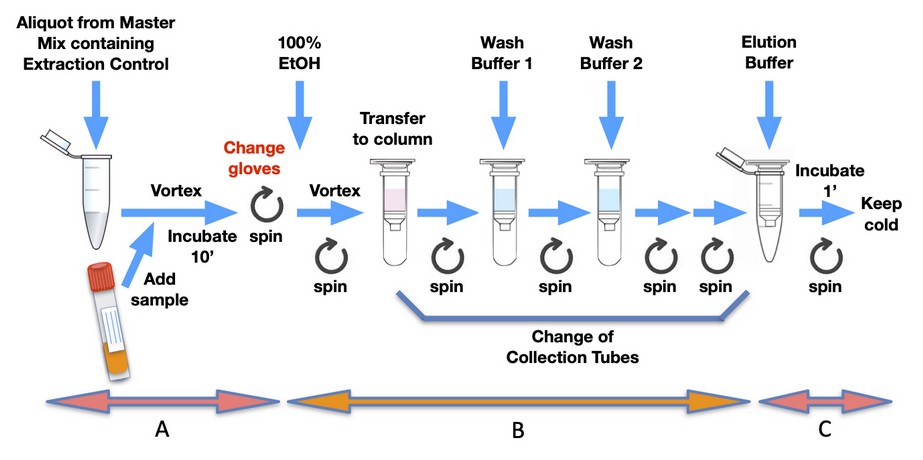
Figure 1. Diagram showing the steps for viral RNA isolation using a standard commercial kit (QIAamp® Viral RNA Mini Kit, Qiagen), with few modifications. Letters A-C at the bottom indicate different types of contamination for the different steps of sample processing mentioned in the text.
The master mix contains an extraction control that helps verify whether the extraction process has been optimal for every individual sample. As an example, this control can be the RNA of a cellular household gene such as actin. This will later appear as a specific curve in the PCR reaction since the primers for that gene will be included in the primer mix used for the PCR reaction. The signal generated during PCR amplification will appear in a different channel than that of the SARS-CoV-2 target gene of interest. The absence of the extraction control curve in all the samples of the batch in the PCR would indicate that it was probably not added to the master mix. If absent in only one or a few samples, it could indicate the presence of inhibitors of the PCR reaction such as ethanol, and the extraction of those particular samples should be repeated.
Once the master mix has been added to all the microcentrifuge tubes, it is time to place the nasopharyngeal swab samples inside the hood, which had been kept in a refrigerator upon arrival. Careless manipulation of swab samples can generate mix-ups leading to false positives. The manipulation of patient samples entails a high risk of contamination (arrow A, bottom of Fig. 1) that can then be passed down throughout the extraction process. Tubes should ideally be opened with a hand that does not hold the micropipette to avoid contaminating it. A brief vortex ensures a good mix for every sample and potentially generates aerosols that can contaminate the working area. That is why it is essential to work under the hood, with the airflow removing those aerosols. Aerosols can also adhere to the shaft of the micropipettes, especially when they are introduced deep into the patient’s sample tubes or touch the swabs that usually come within. If that happens, the parts suspected of being contaminated should be wiped with 70% ethanol between pipetting samples. Once the micropipette tip is loaded with the sample, keeping it at an angle instead of vertically prevents dripping of the content for a sufficiently long time before adding it to the microcentrifuge tube. Also, it is common practice in molecular biology or microbiology labs to eject the residual volume of the tip with an extra push of the micropipette plunger. This, however, is an essential source of aerosols. Therefore, it is desirable to avoid ejecting that residual volume to avoid cross-contamination during the extraction of SARS-CoV-2 viral RNA.
The negative control of the extraction (NCE) must be the last microcentrifuge tube of every batch of samples to ensure that contamination is appropriately detected. Nuclease-free water is added to the tube instead of the viral sample. Being the last tube of the batch, the NCE will help identify cross-contamination events from aerosols of a positive sample of the same batch. In the PCR, the NCE should only yield the curve corresponding to the extraction control. In the case the NCE produced the curve corresponding to the SARS-CoV-2 gene being tested, the extraction of the whole batch should be repeated. Fig. 2A shows the fluorescence profile of the extraction control in the amplification plot of a positive sample.
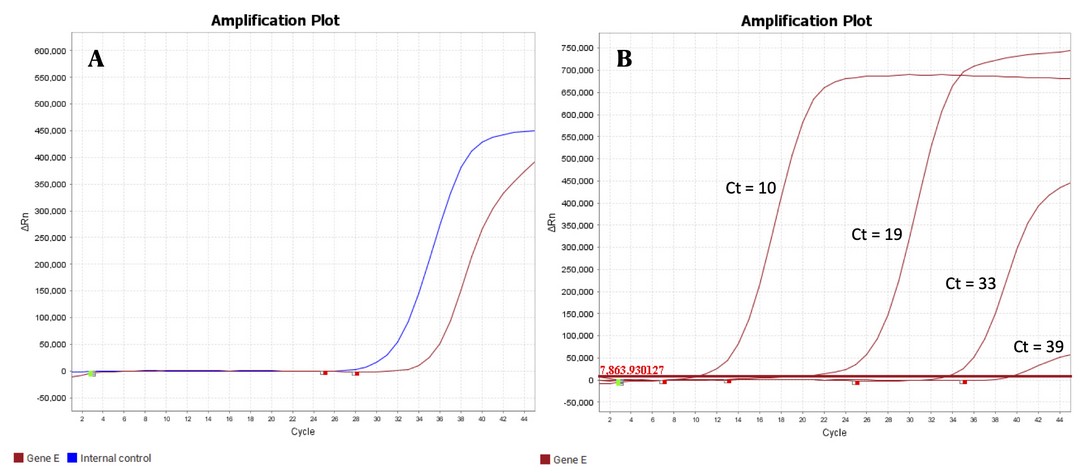
Figure 2. A) Amplification plot of a positive sample showing the extraction control (blue line) and the amplification of the SARS-CoV-2 gene (red line); B) Comparison of the amplification plots of two samples with low Ct (left) and two with high Ct (right). The horizontal line indicates the threshold of the PCR reaction.
Finally, many extraction kits require a few minutes to inactivate the virus. That time can be used to clean surfaces in contact with patient samples, tip boxes, micropipettes, and the microcentrifuge tubes’ exterior. Once the samples have been inactivated in the first steps of the RNA extraction process, the risk of acquiring floating virus particles is significantly reduced, and many labs continue their work on the bench. However, the risk of cross-contamination among samples is higher on the bench than on the laminar flow hood. Hence, although not deemed necessary for the protection of the lab technician, a laminar flow hood helps avoid the generation of false positives among samples. During the intermediate steps of the extraction process, errors during pipetting and handling of samples and reagents account for additional cross-contamination risks (arrow B, bottom of Fig. 1). It is advisable always to assume that aerosols from positive samples are present in the air to increase the sense of alertness and carefulness during the handling of microcentrifuge tubes, buffer solutions, and micropipettes. When using the micropipette, the tips should not enter too deep into solutions because that facilitates the carryover of the solution and its posterior dripping. Very importantly, buffer solutions should be opened only when needed, keeping tubes closed in the meantime.
Most extraction kits require the use of small centrifuges for several steps. The centrifuges are challenging to clean from aerosols generated from positive samples during the manipulation of the tubes. A high risk of contamination occurs in the elution step (arrow C, bottom of Fig. 1) when a column is commonly placed inside an open microcentrifuge tube. Having it open, the lid of the microcentrifuge tube sometimes breaks during the final elution spin. Again, it is essential to use tightly fitting gloves when picking up the tubes and columns from the centrifuge. Centrifuges should be cleaned thoroughly after the processing of every batch of samples.
The use of automatized systems for RNA extraction improves the processing speed enormously and reduces the chances of cross-contamination. During manual feeding of patient samples to the robot in the laminar flow hood, errors can still happen at the initial step of the process. If possible, the operator should close all the wells that are not being used. To avoid pipetting errors, it is advised to mark during the loading process the wells of the plate where the sample has been already added. Once the work under the laminar flow hood has ended, it must be cleaned, and UV irradiated to inactivate any virus particles that could have escaped in aerosols during the extraction process15. Automatized systems also need to be UV irradiated after use.
Reduction of false positives in the pre-PCR area
The most common method for producing reliable test results involves the use of an RT-PCR thermocycler. The conversion of the extracted RNA into DNA is carried out by a retro transcriptase, while a DNA polymerase does further amplification of that DNA. Both enzymes can be part of the same mix (one-step RT-PCR) or separate reactions (two-step RT-PCR). The PCR mix must include primers and fluorescent probes for a SARS-CoV-2 gene and for the extraction RNA target gene that was added during viral RNA extraction as a control. These reagents are added in the pre-PCR area, space physically separated from the room in which the thermocycler is used. In the pre-PCR area, the lab technician will aliquot the PCR mix in the wells of PCR plates or tubes and then add the extracted RNA and controls.
There is absolutely no risk for the lab technicians in the pre-PCR area to be contaminated with active viruses. They can, however, bring contamination to the sample preparation area on her clothes. To avoid carrying over amplicons or viral RNA on their clothes, personnel who work in the pre-PCR area must use a different lab coat and gloves than those used in other areas of the laboratory. Although in principle, the assembling of the PCR can take place on the bench, it is advisable to use a laminar flow hood for that purpose, similar to what was recommended for the extraction area, to prevent aerosols from positive RNA samples contaminating the wells of the PCR strips or plates. In Ecuador, a laminar flow hood is a mandatory requirement for this area. We must never forget how sensitive the PCR technique is and detect as few as 5 copies of viral cDNA4. Similar to what is done in the extraction area, the laminar flow hood must be irradiated for 10-20 minutes with UV light until the airflow stabilizes before assembling the PCR reaction.
Despite working under the laminar flow hood, we must assume that aerosols containing RNA from positive samples could still be present and cause false positives. Hence, a good piece of advice for the assembly of the PCR is to keep all the wells in the PCR strips or plates covered at all times and only lift the caps (strips) or optical film (plates) when a new sample must be added to the appropriate well. Keeping nearby a log of the position of the samples in the wells helps to prevent mistakes during sample loading. Again, to prevent the spread of aerosols, it is essential to use very fit gloves, without folds on the surfaces of the fingertips, to properly open and cap the tubes (strips) or hand the optical film (plates). The closing of the caps of PCR tubes may require applying great force, which can cause vibrations on the cold rack that supports the strips and make them move or fall off the rack. For that reason, it is good to write a small number or another type of code on top of each PCR strip that indicates the orientation and position of wells in the PCR to be run. Another suggestion to avoid aerosols is to not pipet the sample up and down to mix in with the PCR reagents in each tube. This common practice is not needed since the PCR strips or plates will be spun down in a centrifuge before taking them to the thermocycler. Combined with the high temperatures of the PCR process itself, the homogeneity of the reagent solution is assured.
The PCR reaction’s assembly, a negative and one positive control, respectively, needs to be included. The negative control (Non-Template Control, NTC) contains the mix with the buffer, polymerase, and primers for the PCR reaction but nuclease-free water is added instead of extracted RNA. The control also lacks the extraction control added to the samples processed in the RNA extraction area. At the end of the PCR run, the NTC profile should be a flat line for all the channels. The positive control (PC) has the same content as the NTC, but instead of water, the same gene of the SARS-CoV-2 virus that the primers detect in the actual patient samples. The PC also lacks the extraction control added to the samples in the extraction process, and it also emits fluorescence in a different channel. Fig. 2A shows the fluorescence profile from a positive sample for SARS-CoV-2 at the end of the PCR run.
Both NTC and PC must be placed at the end of the assembly of the PCR reaction after all the other samples have been loaded. In this way, the NTC will serve the purpose of letting us know whether cross-contamination has taken place during the assembly process. If the NTC yields a positive signal for the gene of interest, the PCR should be repeated because either the master mix was contaminated or aerosols from a positive sample were spread during the assembly process to other samples. If possible, the negative controls from the RNA extraction step (NCEs) should not be added at the end to the PCR strips or plates to minimize their possible contamination with aerosols from positive samples during the assembly process. In this way, if the NCEs yield a positive signal for the viral gene of interest but the NTC produces a negative signal, we could be confident that the cross-contamination event has taken place during the RNA extraction steps and not in the pre-PCR area. If, on the other hand, one suspects contamination events taking place in the pre-PCR area, it is advisable to aliquot known negative samples or NTCs to the PCR plate and check afterward whether they yield a positive signal.
Although we focus mainly on the events that can generate false-positive results, there is also a chance of generating false negatives in the pre-PCR area. One way is to accidentally not pipetting up any liquid from the RNA sample. This can happen, for example, if there is an air bubble in the microcentrifuge tube and the tip of the micropipette just absorbs air instead of sample. That is why it is important not to lower the concentration during work on the bench and always look at the tips to make sure the liquid is absorbed and again check after centrifugation that every tube in the strip or plate has the same volume. Staying focused and being mindful of every movement of the hands above the wells in the strips or plate helps minimize the risk of contamination at every step of the process. Where available, the use of multichannel pipettes also greatly helps to speed up the PCR assembly process. However, they can also fail to acquire the desired volume in some of the channels and be a source of false negatives.
Reduction of false positives in the PCR area
An essential source of false positives that can originate in the PCR area is amplicon contamination. This occurs when the products of a previous PCR are accidentally released and migrate to other parts of the laboratory. The tubes with positive samples contain trillions of copies of the amplified genes that could easily contaminate future extraction processes and PCR assemblies. Even an initially slightly positive sample with a low viral load whose manipulation would not represent much risk during the extraction process can cause significant problems once the target genes have been exponentially multiplied16. To avoid this type of risk, PCR tubes or plates should be disposed of outside the PCR room at the end of the PCR run. Both the pre-PCR and PCR rooms must be physically separated and have independent air extraction systems. It is recommended not to cross from the PCR room to the extraction and pre-PCR rooms wearing the same protective clothing. Another possible source of false positives occurs when tube strips or plates are not adequately balanced during the centrifugation before the PCR run. This causes vibrations that can disperse the contents of the PCR tubes onto their walls, leading to underestimated readings by the thermocycler.
The best way to prevent amplicon contamination is the implementation of a regular cleaning protocol for all surfaces in the lab-based on 10% (w/v) sodium hypochlorite -a chemical amplicon oxidizer- followed by a rinse with 70% ethanol (to avoid the corrosive effect of the bleach on equipment)17,18. Commercial products abound that include diluted NaOH in their formulations. The use of UV irradiation in Class II biosafety cabinets14 or portable UV lights can also be of great help. Inactivating enzymatic methods such as degradation by uracyl-N-glycosylase19,20 can be included in the PCR reaction mix. On the other hand, inactivation protocols that require the opening of the PCR tubes after amplification are to be avoided.
The Ct (cycle threshold) is the cycle number when the fluorescence of the amplicons surpasses the threshold fluorescence during the PCR run. In our lab, based on the experience of other labs using the same diagnostics kits, a Ct value of 40 or earlier is interpreted as a positive result for COVID-19. The Ct value also measures how many copies of the target gene were in the original sample. The smaller the Ct, the more viral copies the sample has. Fig. 2B shows the amplification plots of four samples, two of them with low Ct values indicating high titers of the target gene. A sample with Ct 10 contains thousands of more copies in origin than a sample of Ct 20. Extraction of samples with high viral titers constitutes a vital source of contamination for the subsequent samples in the batch due to the unintended release of aerosols20. One can never know beforehand which sample has a high viral titer; this is learned after the PCR. Automatic extraction reduces the risk of cross-contamination from high viral titer samples.
In the analysis post-PCR analysis, the lab personnel must decide in every case whether the sample is positive or negative, looking both at the Ct values and the curves. False positives can appear due to the presence of inviable viral particles in patients who are in the process of clearing the remains of the virus from their bodies. If the data look sound, the lab must report those results as positives and let the clinicians make the final decision based on the patients’ medical history, local rates of COVID-19 infections, patient’s signs and symptoms of the disease, or serological analysis6,10,21.
When the signal of the SARS-CoV-2 gene in the PCR appears with a Ct value close to (above or below) 40 (like the sample with Ct 39 in Fig. 2B), a definite diagnosis should not yet be made by the lab processing the samples. Such a profile could result from contamination with aerosols of a close sample with a very low Ct value. The fastest solution is to repeat the extraction of that particular sample on the same day. Alternatively, one can request another sample of the same patient within 2-3 days: if the patient happened to be at the beginning of the infection process, she will appear positive in the second PCR, while a negative result would mean clearance of the virus at the end of the infection period. On average, the best time to get a sample that results in a lower chance of false-negative is eight days after infection or three days after the onset of symptoms1.
All lab members in charge of the analysis post-PCR should be using precisely the same criteria. For that, an algorithm like the one in Fig. 3 can be devised and agreed upon. That way, based on the PCR results, any user can quickly troubleshoot unusual results, find the cause of false positives, and make amendments before reporting the results. Note that despite all the controls being correct at the bottom of Figure 3, one must still be wary of an unusual number of positive samples, especially if they appear grouped. In this case, we recommend that a subset of the samples that also includes negatives be subjected to another round of viral RNA extraction and PCR. As an internal quality control measure, a small fraction of the daily samples arriving at the lab can be randomly selected for retesting. The selection of samples should be made before knowing the results of the PCR test, allowing for the inclusion of both positive and negative samples.
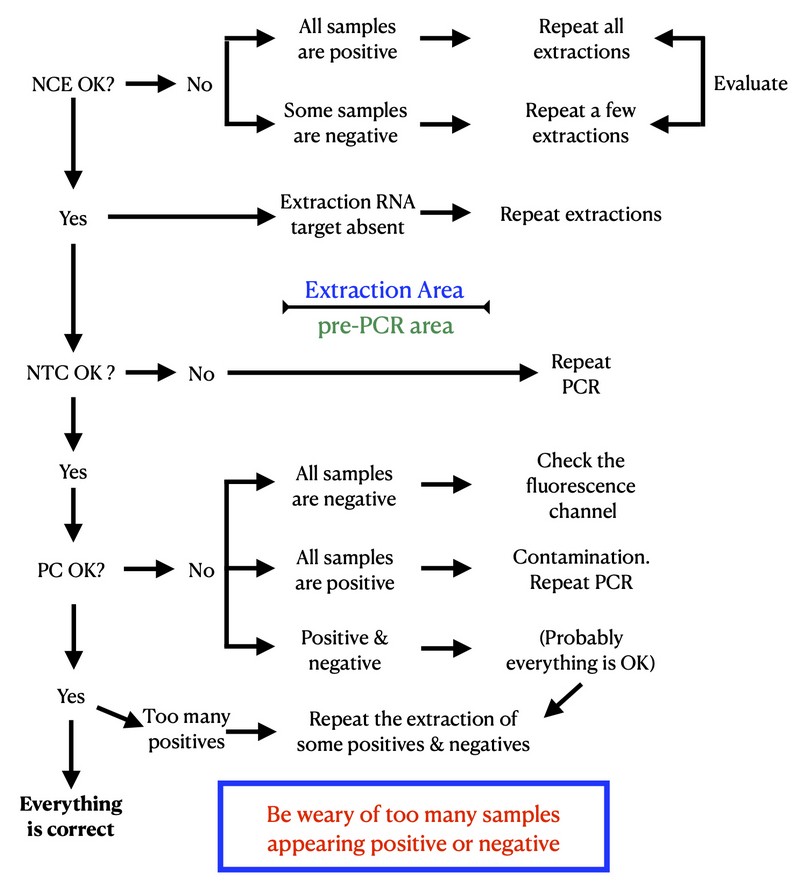
Figure 3. Algorithm post-PCR for the evaluation of the results. NCE, negative control of the extraction; NTC, negative control of the PCR; PC, positive control of the PCR.
DISCUSSION
Having navigated through all the steps of manual sample processing in a laboratory of COVID-19 testing, one must not forget the importance of selecting the best personnel for the different tasks to be performed. Good leadership is necessary to be aware of the strengths and weaknesses of every person working in the lab and get the best out of everyone. False positives are mainly produced through unintentional errors during viral RNA extraction, the assembly of the PCR, and even data management. Hence, a tremendous amount of focus and manual dexterity is expected in laboratory workers to avoid pipetting errors. Maintaining focus is essential in the COVID-19 testing laboratory since many everyday tasks can be repetitive, and the operator may easily engage in detrimental mind-wandering behavior for brief periods of time21. At times the amount of COVID-19 samples arriving at the lab is too tremendous, and several shifts are required to process them. The schedules must be made to avoid too much strain on the workers, thus ensuring that the workers enjoy what they do and maintain a positive attitude in the lab. Remembering that sample numbers are numbers and represent people and their families can be another source of motivation.
It is common practice to interpret PCR results in the context of the pretest probability of the disease13. For example, a patient that appears positive for COVID-19 by PCR but has no symptoms or medical history of the disease has no antibodies and was not exposed to the disease could be considered a false positive. In our opinion, that is a call that doctors should make but not be the labs reporting their results. In case of doubt, it is always better to repeat the RNA extraction or PCR or ask for a new sample. Similarly, a positive result in someone already known to have contracted the disease weeks ago is still positive, although she may not be infectious anymore and is probably just shedding inviable virus particles22.
Samples with low Ct values cause more trouble in the lab in terms of the production of false positives than those of high Ct values during the manual processing of samples. Although sampling introduces a great deal of variability in the first place, low Ct values reflect high titers of the SARS-CoV-2 gene. This seems associated with a high viral load in the original patient samples, for those with low Ct values are more culture-positive than those with high Ct values23 and correlate with the risk of intubation and in-hospital mortality12,24. Hence, it could seem that reporting the Ct value would be very useful for physicians. However, the issue is up for debate nowadays, mainly due to the high Ct values between and within methods11 and during sampling.
CONCLUSIONS
From our experience, it is virtually impossible to eliminate false positives in the COVID-19 diagnostics lab completely. Manual processing requires multiple manipulations of samples and reagents, which translates easily into events of contamination. Automatized systems can significantly reduce (but not eliminate) the appearance of false-positive results. Contamination from samples with very low Ct values is more likely to occur during the extraction process than during the assembly of the PCR. It is advisable to routinely repeat the extraction of positive samples from a batch in which very high viral titer samples were included, especially when too many positives appear clustered together. Also, as an internal quality control measure to gain confidence in their results, labs can temporarily keep a small fraction of their daily samples, chosen randomly, for retesting. With the comments of this article, we hope to contribute to a reduction in the rate of false positives in labs dedicated to similar tests elsewhere during the manual processing of COVID-19 samples.
Declarations
Funding: No funds, grants, or other support was received.
Conflicts of interest/Competing interests: Francisco Alvarez, Mariela Perez, Marco Gudiño, and Markus Tellkamp declare that they have no conflicts of interest.
Ethics approval: Not applicable.
Consent to participate: Not applicable.
Consent for publication: Not applicable.
Availability of data and material: Not applicable.
Code availability: Not applicable.
Authors’ contributions: Francisco J. Alvarez wrote the manuscript and created the figures. Mariela Perez-Cardenas, Marco Gudiño-Gomezjurado and Markus P. Tellkamp edited the manuscript.
Acknowledgments
We want to thank GIZ (Deutsche Gesellschaft für Internationale Zusammenarbeit) for their support, providing equipment, detection kits, and reagents for the majority of tests at Yachay Tech University. We also want to thank the Prefecture of Imbabura, local governments, and enterprises for their help in adapting our university labs for COVID-19 testing.
We also want to acknowledge the rest of the team COVID at Yachay Tech University: Karla Miño, Abigail Montero, Daniela Navas-León, María Paula Romero, Carlos Pazmiño, Karen Sánchez and Alexandra Yépez.
REFERENCES
1. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J (2020) Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann Intern Med 173(4):262-267. https://doi.org/10.7326/M20-1495
2. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W (2020) Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. https://doi.org/10.1001/jama.2020.3786
3. Kipkorir V, Cheruiyot I, Ngure B, Misiani M, Munguti J. Prolonged SARS-CoV-2 RNA detection in anal/rectal swabs and stool specimens in COVID-19 patients after negative conversion in nasopharyngeal RT-PCR test. J Med Virol. 2020 Nov;92(11):2328-2331. doi: 10.1002/jmv.26007. Epub 2020 2 August. PMID: 32401374; PMCID: PMC7272912.
4. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans B, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3):2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045
5. Sethuraman N, Jeremiah SS, Ryo A (2020) Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 323(22):2249–2251. https://doi.org/10.1001/jama.2020.8259
6. Watson J, Whiting PF, Brush JE (2020) Interpreting a covid-19 test result. BMJ. 369:m1808. https://doi.org/10.1136/bmj.m1808
7. Albendín-Iglesias H, Mira-Bleda E, Roura-Piloto AE, Hernández-Torres A, Moral-Escudero E, Fuente-Mora C, Iborra-Bendicho A, Moreno Docón A, Galera Peñaranda C, García Vázquez E (2020) Usefulness of the epidemiological survey and RT-PCR test in pre-surgical patients for assessing the risk of COVID-19. J Hosp Inf 105(4):773-5. https://doi.org/10.1016/j.jhin.2020.06.009
8. Katz AP, Civantos FJ, Sargi Z, Leibowitz JM, Nicolli EA, Weed D, Moskovitz AE, Civantos AM, Andrews DM, Martinez O, Thomas GR (2020) False-positive reverse transcriptase polymerase chain reaction screening for SARS-CoV-2 in the setting of urgent head and neck surgery and otolaryngologic emergencies during the pandemic: Clinical implications. Head & Neck 42:1621-8. https://doi.org/10.1002/hed.26317
9. Cohen AN, Kessel B (2020) False positives in reverse transcription PCR testing for SARS-CoV-2. medRxiv preprint https://doi.org/10.1101/2020.04.26.20080911
10. Cohen AN, Kessel B, Milgroom MG (2020) Diagnosing SARS-CoV-2 infection: the danger of over-reliance on positive test results. medRxiv preprint https://doi.org/10.1101/2020.04.26.20080911
11. Rhoads D, Peaper DR, She RC, Nolte FS, Wojewoda CM, Anderson NW, Pritt BS (2020) College of American Pathologists (CAP) Microbiology Committee Perspective: Caution must be used in interpreting the Cycle Threshold (Ct) value. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa1199
12. Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, Goyal P, Safford MM, Satlin MJ (2020) Impact of Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients With Coronavirus Disease 2019. Clinical Infectious Diseases. https://doi.org/10.1093/cid/ciaa851
13. Surkova E, Nikolayevskyy V, Drobniewski, F (2020) False-positive COVID-19 results: hidden problems and costs. The Lancet. https://doi.org/10.1016/S2213-2600(20)30453-7
14. Padua R, Parrado A, Larghero J, Chomienne C (1999) UV and clean air result in contamination-free PCR. Leukemia 13:1898–1899. https://doi.org/10.1038/sj.leu.2401579
15. Storm N, McKay LGA, Downs SN, Johnson RI, Birru D, de Samber M, Willaert W, Cennini G, Griffiths A (2020). Rapid and complete inactivation of SARS-CoV-2 by ultraviolet-C irradiation. Sci Rep 10, 22421. https://doi.org/10.1038/s41598-020-79600-8
16. Persing DH (1990) Polymerase chain reaction: trenches to benches. J Clin Microbiol 29:1281-1285
17. Hayatsu H, Pan S, Ukita T (1971) Reaction of sodium hypochlorite with nucleic acids and their constituents. Chem Pharm Bull (Tokyo). 19(10):2189-92. https://doi.org/10.1248/cpb.19.2189
18. Kampmann ML, Børsting C, Morling N (2017) Decrease DNA contamination in the laboratories. Forensic Science International: Genetics Supplement Series 6:e577-e578. https://doi.org/10.1016/j.fsigss.2017.09.223
19. Longo MC, Berninger MS, Hartley JL (1990) Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93(1):125-8. https://doi.org/10.1016/0378-1119(90)90145-h
20. Sefers S, Stratton CW, Tang YW (2013) Amplification Product Inactivation. In: Tang YW., Stratton C. (eds) Advanced Techniques in Diagnostic Microbiology. Springer, Boston, MA. https://doi.org/10.1007/978-1-4614-3970-7_26
21. McVay, J. C., & Kane, M. J. (2012). Drifting from slow to «d’oh!»: Working memory capacity and mind wandering predict extreme reaction times and executive control errors. Journal of Experimental Psychology: Learning, Memory, and Cognition, 38(3), 525–549. doi:10.1037/a0025896
22. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C (2020) Virological assessment of hospitalized patients with COVID-2019. Nature. https://doi.org/10.1038/s41586-020-2196-x
23. Jaafar R, Aherfi S, Wurtz N, Grimaldier C, Van Hoang T, Colson P, Raoult D, La Scola B (2020) Correlation Between 3790 Quantitative Polymerase Chain Reaction–Positives Samples and Positive Cell Cultures, Including 1941 Severe Acute Respiratory Syndrome Coronavirus 2 Isolates. Clinical Infectious Diseases. https://doi.org/10.1093/cid/ciaa1491
24. Rao SN, Manissero D, Steele VR, Pareja J (2020) A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect Dis Ther. 9(3):573-586. https://doi.org/10.1007/s40121-020-00324-3. Erratum in: Infect Dis Ther. PMID: 32725536
Received: 10 February 2021
Accepted: 8 March 2021
Francisco J. Alvarez, Mariela Perez-Cardenas, Marco Gudiño, Markus P. Tellkamp
Francisco Javier Álvarez Botas (Francisco J. Alvarez, corresponding author), ORCID: 0000-0003-1632-1792
Mariela Alexandra Pérez Cárdenas (Mariela Perez-Cardenas)
Marco Esteban Gudiño Gomezjurado (Marco Gudiño), ORCID: 0000-0002-8609-0806
Markus Patricio Tellkamp Tietz (Markus P. Tellkamp), ORCID: 0000-0002-6442-6647
Affiliation
Yachay Tech University, School of Biological Sciences and Engineering, Hda. San José s/n y Proyecto Yachay, 100119, Urcuquí, Ecuador
E-mail
corresponding author. falvarez@yachaytech.edu.ec



















